+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4yxb | ||||||
|---|---|---|---|---|---|---|---|
| Title | FliM(SPOA)::FliN fusion protein | ||||||
 Components Components |
| ||||||
 Keywords Keywords | PROTEIN TRANSPORT / Type III Secretion System | ||||||
| Function / homology |  Function and homology information Function and homology informationbacterial-type flagellum basal body / bacterial-type flagellum-dependent swarming motility / positive chemotaxis / cytoskeletal motor activity / bacterial-type flagellum-dependent cell motility / chemotaxis / plasma membrane Similarity search - Function | ||||||
| Biological species |  Salmonella typhimurium (bacteria) Salmonella typhimurium (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium str. 798 (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium str. 798 (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  SAD / Resolution: 2.56 Å SAD / Resolution: 2.56 Å | ||||||
 Authors Authors | Notti, R.Q. / Stebbins, C.E. | ||||||
 Citation Citation |  Journal: Nat Commun / Year: 2015 Journal: Nat Commun / Year: 2015Title: A common assembly module in injectisome and flagellar type III secretion sorting platforms. Authors: Notti, R.Q. / Bhattacharya, S. / Lilic, M. / Stebbins, C.E. | ||||||
| History |
|
- Structure visualization
Structure visualization
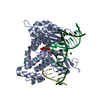
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4yxb.cif.gz 4yxb.cif.gz | 83.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4yxb.ent.gz pdb4yxb.ent.gz | 60.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4yxb.json.gz 4yxb.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/yx/4yxb https://data.pdbj.org/pub/pdb/validation_reports/yx/4yxb ftp://data.pdbj.org/pub/pdb/validation_reports/yx/4yxb ftp://data.pdbj.org/pub/pdb/validation_reports/yx/4yxb | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 2 types, 3 molecules ABD
| #1: Protein | Mass: 25085.904 Da / Num. of mol.: 2 Fragment: UNP Residues 245-334,UNP Residues 5-137,UNP Residues 245-334,UNP Residues 5-137 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria) Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria)Strain: LT2 / SGSC1412 / ATCC 700720 Gene: fliM, cheC2, fla AII, fla QII, STM1976, fliN, flaN, motD, STM1977 Production host:  #3: Protein | | Mass: 8911.981 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Salmonella enterica subsp. enterica serovar Typhimurium str. 798 (bacteria), (gene. exp.) Salmonella enterica subsp. enterica serovar Typhimurium str. 798 (bacteria), (gene. exp.)  Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria) Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria)Gene: fliN, UMN798_2086, fliN, flaN, motD, STM1977 / Strain: LT2 / SGSC1412 / ATCC 700720 / Production host:  |
|---|
-Protein/peptide , 2 types, 2 molecules CE
| #2: Protein/peptide | Mass: 690.787 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium (bacteria)Production host:  |
|---|---|
| #4: Protein/peptide | Mass: 528.644 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: This peptide fragment was built into a region of electron density for which the sequence could not be definitively determined. Source: (gene. exp.)  Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium (bacteria)Production host:  |
-Non-polymers , 2 types, 24 molecules 


| #5: Chemical | ChemComp-IMD / |
|---|---|
| #6: Water | ChemComp-HOH / |
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.28 Å3/Da / Density % sol: 46.15 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop Details: FliM(245-334)::FliN(5-137) was concentrated to 7.5mg/mL and crystallized with 2.2M NaCl, 100mM imidazole.Cl pH=8.0. Crystals were cryoprotected with 2M NaCl, 100mM imidazole.Cl pH=8.0, 30% glycerol. |
-Data collection
| Diffraction | Mean temperature: 100 K | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  NSLS NSLS  / Beamline: X29A / Wavelength: 0.979 Å / Beamline: X29A / Wavelength: 0.979 Å | |||||||||||||||||||||||||||
| Detector | Type: ADSC QUANTUM 315r / Detector: CCD / Date: Sep 23, 2014 | |||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 0.979 Å / Relative weight: 1 | |||||||||||||||||||||||||||
| Reflection | Resolution: 2.56→57.67 Å / Num. obs: 18420 / % possible obs: 100 % / Redundancy: 13.8 % / Biso Wilson estimate: 66.34 Å2 / CC1/2: 0.999 / Rmerge(I) obs: 0.097 / Rpim(I) all: 0.027 / Net I/σ(I): 18.5 / Num. measured all: 254307 / Scaling rejects: 12 | |||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1 / Rejects: _
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  SAD / Resolution: 2.56→47.078 Å / SU ML: 0.41 / Cross valid method: FREE R-VALUE / σ(F): 1.35 / Phase error: 30.47 / Stereochemistry target values: MLHL SAD / Resolution: 2.56→47.078 Å / SU ML: 0.41 / Cross valid method: FREE R-VALUE / σ(F): 1.35 / Phase error: 30.47 / Stereochemistry target values: MLHL
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 131.14 Å2 / Biso mean: 68.1747 Å2 / Biso min: 11.58 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 2.56→47.078 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 13
|
 Movie
Movie Controller
Controller

















 PDBj
PDBj





