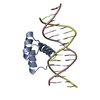
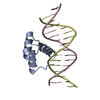
Entry Database : PDB / ID : 4xidTitle AntpHD with 15bp DNA duplex DNA (5'-D(*AP*GP*AP*AP*AP*GP*CP*CP*AP*TP*TP*AP*GP*AP*G)-3')DNA (5'-D(*TP*CP*TP*CP*TP*AP*AP*TP*GP*GP*CP*TP*TP*TP*C)-3')Homeotic protein antennapedia Keywords / / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Drosophila melanogaster (fruit fly)unidentified (others) Method / / / Resolution : 2.701 Å Authors White, M.A. / Zandarashvili, L. / Iwahara, J. Journal : Biophys.J. / Year : 2015Title : Entropic Enhancement of Protein-DNA Affinity by Oxygen-to-Sulfur Substitution in DNA Phosphate.Authors : Zandarashvili, L. / Nguyen, D. / Anderson, K.M. / White, M.A. / Gorenstein, D.G. / Iwahara, J. History Deposition Jan 6, 2015 Deposition site / Processing site Revision 1.0 Nov 25, 2015 Provider / Type Revision 1.1 Mar 16, 2016 Group Revision 1.2 Nov 22, 2017 Group / Refinement description / Category / softwareItem / _software.classificationRevision 1.3 Sep 27, 2023 Group Data collection / Database references ... Data collection / Database references / Derived calculations / Refinement description Category chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / pdbx_struct_conn_angle / struct_conn Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_struct_conn_angle.ptnr1_auth_asym_id / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr2_auth_asym_id / _pdbx_struct_conn_angle.ptnr2_label_asym_id / _pdbx_struct_conn_angle.ptnr2_symmetry / _pdbx_struct_conn_angle.ptnr3_auth_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.value / _struct_conn.pdbx_dist_value / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_symmetry
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT /
MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 2.701 Å
molecular replacement / Resolution: 2.701 Å  Authors
Authors Citation
Citation Journal: Biophys.J. / Year: 2015
Journal: Biophys.J. / Year: 2015 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 4xid.cif.gz
4xid.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb4xid.ent.gz
pdb4xid.ent.gz PDB format
PDB format 4xid.json.gz
4xid.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/xi/4xid
https://data.pdbj.org/pub/pdb/validation_reports/xi/4xid ftp://data.pdbj.org/pub/pdb/validation_reports/xi/4xid
ftp://data.pdbj.org/pub/pdb/validation_reports/xi/4xid
 Links
Links Assembly
Assembly


 Components
Components






 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation ROTATING ANODE / Type: RIGAKU FR-E+ DW / Wavelength: 1.5418 Å
ROTATING ANODE / Type: RIGAKU FR-E+ DW / Wavelength: 1.5418 Å molecular replacement
molecular replacement Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller












 PDBj
PDBj







































