[English] 日本語
 Yorodumi
Yorodumi- PDB-4v57: Crystal structure of the bacterial ribosome from Escherichia coli... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4v57 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of the bacterial ribosome from Escherichia coli in complex with spectinomycin and neomycin. | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | RIBOSOME / RNA-protein complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationstringent response / ornithine decarboxylase inhibitor activity / transcription antitermination factor activity, RNA binding / misfolded RNA binding / Group I intron splicing / RNA folding / transcriptional attenuation / endoribonuclease inhibitor activity / positive regulation of ribosome biogenesis / RNA-binding transcription regulator activity ...stringent response / ornithine decarboxylase inhibitor activity / transcription antitermination factor activity, RNA binding / misfolded RNA binding / Group I intron splicing / RNA folding / transcriptional attenuation / endoribonuclease inhibitor activity / positive regulation of ribosome biogenesis / RNA-binding transcription regulator activity / translational termination / negative regulation of cytoplasmic translation / four-way junction DNA binding / DnaA-L2 complex / translation repressor activity / negative regulation of translational initiation / regulation of mRNA stability / negative regulation of DNA-templated DNA replication initiation / mRNA regulatory element binding translation repressor activity / positive regulation of RNA splicing / assembly of large subunit precursor of preribosome / cytosolic ribosome assembly / response to reactive oxygen species / regulation of DNA-templated transcription elongation / ribosome assembly / transcription elongation factor complex / transcription antitermination / DNA endonuclease activity / regulation of cell growth / DNA-templated transcription termination / response to radiation / maintenance of translational fidelity / mRNA 5'-UTR binding / regulation of translation / large ribosomal subunit / ribosome biogenesis / transferase activity / ribosome binding / ribosomal small subunit biogenesis / ribosomal small subunit assembly / 5S rRNA binding / ribosomal large subunit assembly / small ribosomal subunit / small ribosomal subunit rRNA binding / large ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / cytosolic large ribosomal subunit / cytoplasmic translation / tRNA binding / negative regulation of translation / rRNA binding / structural constituent of ribosome / ribosome / translation / response to antibiotic / negative regulation of DNA-templated transcription / mRNA binding / DNA binding / RNA binding / zinc ion binding / membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 3.5 Å molecular replacement / Resolution: 3.5 Å | |||||||||
 Authors Authors | Borovinskaya, M.A. / Shoji, S. / Holton, J.M. / Fredrick, K. / Cate, J.H.D. | |||||||||
 Citation Citation |  Journal: Acs Chem.Biol. / Year: 2007 Journal: Acs Chem.Biol. / Year: 2007Title: A steric block in translation caused by the antibiotic spectinomycin. Authors: Borovinskaya, M.A. / Shoji, S. / Holton, J.M. / Fredrick, K. / Cate, J.H. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4v57.cif.gz 4v57.cif.gz | 6.3 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4v57.ent.gz pdb4v57.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  4v57.json.gz 4v57.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/v5/4v57 https://data.pdbj.org/pub/pdb/validation_reports/v5/4v57 ftp://data.pdbj.org/pub/pdb/validation_reports/v5/4v57 ftp://data.pdbj.org/pub/pdb/validation_reports/v5/4v57 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4v56C  2avy  2aw4  2aw7  2awb S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
-RNA chain , 3 types, 6 molecules AACABADABBDB
| #1: RNA chain | Mass: 499690.031 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  #22: RNA chain | Mass: 38790.090 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  #23: RNA chain | Mass: 941612.375 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  |
|---|
-30S ribosomal protein ... , 20 types, 40 molecules ACCCADCDAECEAFCFAGCGAHCHAICIAJCJAKCKALCLAMCMAPCPAQCQARCRASCS...
| #2: Protein | Mass: 25900.117 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  #3: Protein | Mass: 23383.002 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  #4: Protein | Mass: 17498.203 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  #5: Protein | Mass: 15727.512 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  #6: Protein | Mass: 19923.959 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  #7: Protein | Mass: 14015.361 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  #8: Protein | Mass: 14755.074 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  #9: Protein | Mass: 11755.597 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  #10: Protein | Mass: 13739.778 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  #11: Protein | Mass: 13636.961 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  #12: Protein | Mass: 12997.271 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  #13: Protein | Mass: 9207.572 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  #14: Protein | Mass: 9593.296 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  #15: Protein | Mass: 8874.276 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  #16: Protein | Mass: 10324.160 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  #17: Protein | Mass: 9577.268 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  #18: Protein | Mass: 26650.475 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  #19: Protein | Mass: 8392.844 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  #20: Protein | Mass: 10290.816 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  #21: Protein | Mass: 11475.364 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  |
|---|
+50S ribosomal protein ... , 29 types, 58 molecules BIDIBCDCBDDDBKDKBPDPBEDEBYDYB0D0B4D4B1D1B3D3BVDVB2D2BLDLBMDM...
-Non-polymers , 5 types, 1961 molecules 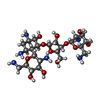

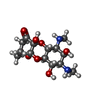






| #53: Chemical | ChemComp-NMY / #54: Chemical | ChemComp-MG / #55: Chemical | #56: Chemical | #57: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 6 X-RAY DIFFRACTION / Number of used crystals: 6 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.41 Å3/Da / Density % sol: 63.98 % | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 277 K / Method: batch / pH: 7.5 Details: MPD, PEG 8000, MgCl2, NH4Cl, spermine, spermidine, TRIS, EDTA, pH 7.5, batch, temperature 277K | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions |
|
-Data collection
| Diffraction | Mean temperature: 93 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ALS ALS  / Beamline: 12.3.1 / Wavelength: 1.111 Å / Beamline: 12.3.1 / Wavelength: 1.111 Å |
| Detector | Type: ADSC QUANTUM 315 / Detector: CCD / Date: Mar 5, 2006 |
| Radiation | Monochromator: Double crystal / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.111 Å / Relative weight: 1 |
| Reflection | Resolution: 3.5→182 Å / Num. obs: 521699 / % possible obs: 72.1 % / Redundancy: 3.3 % / Rmerge(I) obs: 0.104 / Net I/σ(I): 8.6 |
| Reflection shell | Resolution: 3.5→3.85 Å / Redundancy: 1.1 % / Rmerge(I) obs: 0.41 / Mean I/σ(I) obs: 1.9 / % possible all: 13.9 |
-Phasing
| Phasing | Method:  molecular replacement molecular replacement |
|---|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 2AVY, 2AW4, 2AW7, 2AWB Resolution: 3.5→70 Å / σ(F): 0 / Stereochemistry target values: torsional dynamics
| ||||||||||||||||||||||||
| Displacement parameters | Biso mean: 74.9 Å2 | ||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3.5→70 Å
| ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller






















 PDBj
PDBj

































