[English] 日本語
 Yorodumi
Yorodumi- PDB-4uua: Crystal structure of zebrafish Sirtuin 5 in complex with 3S-Z-ami... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4uua | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of zebrafish Sirtuin 5 in complex with 3S-Z-amino- succinylated CPS1-peptide | ||||||
 Components Components |
| ||||||
 Keywords Keywords | HYDROLASE / REGULATORY ENZYME / DEACYLASE / MITOCHONDRIAL / ROSSMANN-FOLD / ZINC-BINDING | ||||||
| Function / homology |  Function and homology information Function and homology informationcarbamoyl phosphate biosynthetic process / cellular response to oleic acid / Transcriptional activation of mitochondrial biogenesis / monoatomic anion homeostasis / regulation of ketone biosynthetic process / peptidyl-lysine demalonylation / protein desuccinylation / peptidyl-lysine desuccinylation / protein-glutaryllysine deglutarylase activity / modified amino acid binding ...carbamoyl phosphate biosynthetic process / cellular response to oleic acid / Transcriptional activation of mitochondrial biogenesis / monoatomic anion homeostasis / regulation of ketone biosynthetic process / peptidyl-lysine demalonylation / protein desuccinylation / peptidyl-lysine desuccinylation / protein-glutaryllysine deglutarylase activity / modified amino acid binding / protein-malonyllysine demalonylase activity / protein-succinyllysine desuccinylase activity / carbamoyl-phosphate synthase (ammonia) activity / triglyceride catabolic process / carbamoyl-phosphate synthase (ammonia) / carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity / midgut development / homocysteine metabolic process / Urea cycle / cellular response to ammonium ion / citrulline biosynthetic process / urea cycle / hepatocyte differentiation / histone deacetylase activity, NAD-dependent / glutamine metabolic process / response to growth hormone / heterocyclic compound binding / glutamate binding / response to zinc ion / response to food / response to starvation / response to dexamethasone / response to amine / small molecule binding / acyl binding / potassium ion binding / mitochondrial nucleoid / NAD+ binding / response to amino acid / 'de novo' pyrimidine nucleobase biosynthetic process / nitric oxide metabolic process / cellular response to fibroblast growth factor stimulus / Transferases; Acyltransferases; Transferring groups other than aminoacyl groups / cellular response to glucagon stimulus / cellular response to cAMP / phospholipid binding / response to toxic substance / vasodilation / response to lipopolysaccharide / endopeptidase activity / mitochondrial inner membrane / mitochondrial matrix / response to xenobiotic stimulus / calcium ion binding / protein-containing complex binding / nucleolus / protein-containing complex / mitochondrion / zinc ion binding / ATP binding / metal ion binding / nucleus / plasma membrane / cytosol / cytoplasm Similarity search - Function | ||||||
| Biological species |   HOMO SAPIENS (human) HOMO SAPIENS (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.8 Å MOLECULAR REPLACEMENT / Resolution: 2.8 Å | ||||||
 Authors Authors | Pannek, M. / Gertz, M. / Steegborn, C. | ||||||
 Citation Citation |  Journal: Angew.Chem.Int.Ed.Engl. / Year: 2014 Journal: Angew.Chem.Int.Ed.Engl. / Year: 2014Title: Chemical Probing of the Human Sirtuin 5 Active Site Reveals its Substrate Acyl Specificity and Peptide-Based Inhibitors. Authors: Roessler, C. / Nowak, T. / Pannek, M. / Gertz, M. / Nguyen, G.T. / Scharfe, M. / Born, I. / Sippl, W. / Steegborn, C. / Schutkowski, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4uua.cif.gz 4uua.cif.gz | 226.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4uua.ent.gz pdb4uua.ent.gz | 182.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4uua.json.gz 4uua.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  4uua_validation.pdf.gz 4uua_validation.pdf.gz | 772.1 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  4uua_full_validation.pdf.gz 4uua_full_validation.pdf.gz | 778.6 KB | Display | |
| Data in XML |  4uua_validation.xml.gz 4uua_validation.xml.gz | 22.2 KB | Display | |
| Data in CIF |  4uua_validation.cif.gz 4uua_validation.cif.gz | 30.3 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/uu/4uua https://data.pdbj.org/pub/pdb/validation_reports/uu/4uua ftp://data.pdbj.org/pub/pdb/validation_reports/uu/4uua ftp://data.pdbj.org/pub/pdb/validation_reports/uu/4uua | HTTPS FTP |
-Related structure data
| Related structure data |  4utnC 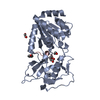 4utrC  4utvC  4utxC  4utzC  4uu7C  4uu8C  4uubC  2nyrS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
| ||||||||
| Noncrystallographic symmetry (NCS) | NCS oper: (Code: given Matrix: (-0.74986, -0.54483, 0.37533), Vector: |
- Components
Components
-Protein / Protein/peptide , 2 types, 3 molecules ABD
| #1: Protein | Mass: 30423.785 Da / Num. of mol.: 2 / Fragment: CATALYTIC CORE, UNP RESIDUES 30-298 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   References: UniProt: Q6DHI5, Hydrolases; Acting on carbon-nitrogen bonds, other than peptide bonds; In linear amides #2: Protein/peptide | | Mass: 969.112 Da / Num. of mol.: 1 / Fragment: UNP RESIDUES 524-531 / Source method: obtained synthetically Details: BENZOYLATED GLYCINE AT POSITION 1 AND 3S-Z-AMINO-SUCCINYL-LYSINE AT POSITION 4 Source: (synth.)  HOMO SAPIENS (human) / References: UniProt: Q5R209, UniProt: P31327*PLUS HOMO SAPIENS (human) / References: UniProt: Q5R209, UniProt: P31327*PLUS |
|---|
-Non-polymers , 7 types, 61 molecules 




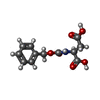







| #3: Chemical | | #4: Chemical | #5: Chemical | ChemComp-EPE / | #6: Chemical | ChemComp-DMS / | #7: Chemical | ChemComp-NA / | #8: Chemical | ChemComp-NX6 / | #9: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.8 Å3/Da / Density % sol: 57 % / Description: NONE |
|---|---|
| Crystal grow | pH: 7.6 / Details: 20% PEG3350, 0.1 M HEPES PH 7.6 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  BESSY BESSY  / Beamline: 14.1 / Wavelength: 0.918409 / Beamline: 14.1 / Wavelength: 0.918409 |
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Apr 17, 2013 / Details: MIRRORS |
| Radiation | Monochromator: SI-111 CRYSTAL / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.918409 Å / Relative weight: 1 |
| Reflection | Resolution: 2.8→50 Å / Num. obs: 18292 / % possible obs: 99.6 % / Observed criterion σ(I): 1.5 / Redundancy: 10.5 % / Rmerge(I) obs: 0.25 / Net I/σ(I): 10.6 |
| Reflection shell | Resolution: 2.8→2.9 Å / Redundancy: 11.1 % / Mean I/σ(I) obs: 1.3 / % possible all: 100 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 2NYR Resolution: 2.8→19.87 Å / Cor.coef. Fo:Fc: 0.946 / Cor.coef. Fo:Fc free: 0.907 / SU B: 40.475 / SU ML: 0.369 / Cross valid method: THROUGHOUT / ESU R: 2.357 / ESU R Free: 0.37 / Stereochemistry target values: MAXIMUM LIKELIHOOD Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS. U VALUES WITH TLS ADDED. N-TERMINAL RESIDUES OF PROTEIN CHAINS ARE DISORDERED. RESIDUES A280 TO A281 ARE DISORDERED. THE LYSINE- ...Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS. U VALUES WITH TLS ADDED. N-TERMINAL RESIDUES OF PROTEIN CHAINS ARE DISORDERED. RESIDUES A280 TO A281 ARE DISORDERED. THE LYSINE-MODIFICATION PRESENT IN THIS COMPLEX STRUCTURE IS AN ENANTIOMER. THE OTHER ENANTIOMERIC FORM DID NOT BIND AND INHIBIT THE ENZYME AS GOOD AS THIS ONE.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 70.386 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.8→19.87 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj











