[English] 日本語
 Yorodumi
Yorodumi- PDB-4buh: Human IgE against the major allergen Bet v 1 - Crystal structure ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4buh | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human IgE against the major allergen Bet v 1 - Crystal structure of clone M0418 scFv | |||||||||
 Components Components | CLONE M0418 SCFV | |||||||||
 Keywords Keywords | IMMUNE SYSTEM / ANTIBODY / ALLERGEN | |||||||||
| Function / homology | Immunoglobulins / Immunoglobulin-like / Sandwich / Mainly Beta / DI(HYDROXYETHYL)ETHER Function and homology information Function and homology information | |||||||||
| Biological species |  HOMO SAPIENS (human) HOMO SAPIENS (human) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.3 Å MOLECULAR REPLACEMENT / Resolution: 1.3 Å | |||||||||
 Authors Authors | Davies, A.M. / Levin, M. / Lilljekvist, M. / Carlsson, F. / Gould, H.J. / Sutton, B.J. / Ohlin, M. | |||||||||
 Citation Citation |  Journal: Clin.Exp.Allergy / Year: 2014 Journal: Clin.Exp.Allergy / Year: 2014Title: Human Ige Against the Major Allergen Bet V 1 - Defining an Epitope with Limited Cross-Reactivity between Different Pr-10 Family Proteins Authors: Levin, M. / Davies, A.M. / Lilljekvist, M. / Carlsson, F. / Gould, H.J. / Sutton, B.J. / Ohlin, M. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4buh.cif.gz 4buh.cif.gz | 213.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4buh.ent.gz pdb4buh.ent.gz | 172.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4buh.json.gz 4buh.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/bu/4buh https://data.pdbj.org/pub/pdb/validation_reports/bu/4buh ftp://data.pdbj.org/pub/pdb/validation_reports/bu/4buh ftp://data.pdbj.org/pub/pdb/validation_reports/bu/4buh | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2yc1S S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 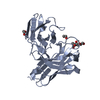
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
| ||||||||
| Noncrystallographic symmetry (NCS) | NCS oper: (Code: given Matrix: (0.51519, -0.08823, 0.85252), Vector: |
- Components
Components
| #1: Antibody | Mass: 28715.406 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  HOMO SAPIENS (human) / Production host: HOMO SAPIENS (human) / Production host:  #2: Chemical | ChemComp-EDO / #3: Chemical | ChemComp-PEG / | #4: Water | ChemComp-HOH / | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.05 Å3/Da / Density % sol: 39.99 % / Description: NONE |
|---|---|
| Crystal grow | pH: 7.5 / Details: 0.1M MES PH6.5, 25% PEG 4000, pH 7.5 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I02 / Wavelength: 0.9795 / Beamline: I02 / Wavelength: 0.9795 |
| Detector | Type: ADSC CCD / Detector: CCD / Date: Mar 18, 2012 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9795 Å / Relative weight: 1 |
| Reflection | Resolution: 1.3→27.3 Å / Num. obs: 109350 / % possible obs: 96.1 % / Observed criterion σ(I): 0 / Redundancy: 9.1 % / Biso Wilson estimate: 12.58 Å2 / Rmerge(I) obs: 0.11 / Net I/σ(I): 12.6 |
| Reflection shell | Resolution: 1.3→1.37 Å / Redundancy: 7.8 % / Rmerge(I) obs: 0.87 / Mean I/σ(I) obs: 3.5 / % possible all: 93.7 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 2YC1 Resolution: 1.3→27.283 Å / SU ML: 0.11 / σ(F): 0 / Phase error: 15.91 / Stereochemistry target values: ML Details: SCFV CONSTRUCT IS NUMBERED SEQUENTIALLY.NUMBERS IN THE PUBLICATION ASSOCIATED WITH THIS ENTRY USE THE IMGT STANDARD NUMBERING SYSTEM. IN BOTH MOLECULES, TRP47 ADOPTS AN ALTERNATIVE ...Details: SCFV CONSTRUCT IS NUMBERED SEQUENTIALLY.NUMBERS IN THE PUBLICATION ASSOCIATED WITH THIS ENTRY USE THE IMGT STANDARD NUMBERING SYSTEM. IN BOTH MOLECULES, TRP47 ADOPTS AN ALTERNATIVE CONFORMATION. ONE CONFORMER AT 0.5 OCCUPANCY FORMS A HYDROGEN BOND WITH A WATER MOLECULE AT 0.5 OCCUPANCY,WHICH OCCUPIES PART OF THE SAME SPACE AS THE SECOND TRP47 CONFORMER.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.3→27.283 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj






