+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1ap2 | ||||||
|---|---|---|---|---|---|---|---|
| Title | SINGLE CHAIN FV OF C219 | ||||||
 Components Components | (MONOCLONAL ANTIBODY C219) x 2 | ||||||
 Keywords Keywords | IMMUNOGLOBULIN / SINGLE CHAIN FV / MONOCLONAL ANTIBODY / C219 / P-GLYCOPROTEIN | ||||||
| Function / homology | Immunoglobulins / Immunoglobulin-like / Sandwich / Mainly Beta / : / :  Function and homology information Function and homology information | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.36 Å MOLECULAR REPLACEMENT / Resolution: 2.36 Å | ||||||
 Authors Authors | Hoedemaeker, P.J. / Rose, D.R. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 1997 Journal: J.Biol.Chem. / Year: 1997Title: A single chain Fv fragment of P-glycoprotein-specific monoclonal antibody C219. Design, expression, and crystal structure at 2.4 A resolution. Authors: Hoedemaeker, F.J. / Signorelli, T. / Johns, K. / Kuntz, D.A. / Rose, D.R. | ||||||
| History |
|
- Structure visualization
Structure visualization
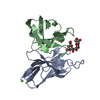
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1ap2.cif.gz 1ap2.cif.gz | 100.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1ap2.ent.gz pdb1ap2.ent.gz | 77.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1ap2.json.gz 1ap2.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ap/1ap2 https://data.pdbj.org/pub/pdb/validation_reports/ap/1ap2 ftp://data.pdbj.org/pub/pdb/validation_reports/ap/1ap2 ftp://data.pdbj.org/pub/pdb/validation_reports/ap/1ap2 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1igmS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||
| 2 | 
| ||||||||||||
| Unit cell |
| ||||||||||||
| Noncrystallographic symmetry (NCS) | NCS oper:
|
- Components
Components
| #1: Antibody | Mass: 12296.612 Da / Num. of mol.: 2 / Fragment: FV Source method: isolated from a genetically manipulated source Details: LIGHT AND HEAVY CHAINS LINKED WITH A SYNTHETIC (GGGGS)3 LINKER Source: (gene. exp.)   #2: Antibody | Mass: 13572.992 Da / Num. of mol.: 2 / Fragment: FV Source method: isolated from a genetically manipulated source Details: LIGHT AND HEAVY CHAINS LINKED WITH A SYNTHETIC (GGGGS)3 LINKER Source: (gene. exp.)   #3: Water | ChemComp-HOH / | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.85 Å3/Da / Density % sol: 58 % | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Method: vapor diffusion, hanging drop / pH: 4.5 Details: THE PROTEIN WAS CRYSTALLIZED IN 21% PEG 6000, 100 MM SODIUM CITRATE PH 4.5, IN HANGING DROPS CONTAINING SUBTILISIN CARLSBERG IN A 1:100 MOLAR RATIO., vapor diffusion - hanging drop THE ...Details: THE PROTEIN WAS CRYSTALLIZED IN 21% PEG 6000, 100 MM SODIUM CITRATE PH 4.5, IN HANGING DROPS CONTAINING SUBTILISIN CARLSBERG IN A 1:100 MOLAR RATIO., vapor diffusion - hanging drop THE DEPOSITORS CRYSTALLIZED THE SINGLE CHAIN FV IN THE PRESENCE OF SUBTILISIN, AS DESCRIBED IN THE PRIMARY REFERENCE. THE SEQUENCE OF THE SINGLE CHAIN IS DIVMTQSPSSLTVTAGEKVTMSCKSSQSLLNSGNQKNYLTWYQQKPGQPPKLLIYWAS TRESGVPDRFTGSGSGTDFTLTISSVQAEDLAVYYCQNDYSYPLTFGAGTKLEP (LIGHT CHAINS A AND C) GGGGSGGGGSGKSGGGG (LINKER) EVQLQQSGAELVRPGASVKLSCTASGFNIKDDFMHWVKQRPEQGLEWIGRIDPANDNT KYAPKFQDKATIIADTSSNTAYLQLSSLTSEDTAVYYCARREVYSYYSPLDVWGAGTT VTVPSG (HEAVY CHAINS B AND D) SEQKLISEEDLNHHHHH (C-MYC TAG + 5XHIS TAG) THE DEPOSITORS HAVE NOT DETERMINED WHERE SUBTILISIN CLEAVES EXACTLY, BUT THE MOBILITY ON SDS GELS SUGGESTS THAT THE MOST OF THE LINKER AND MOST OF BOTH TAGS ARE REMOVED. | ||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 298 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU / Wavelength: 1.5418 ROTATING ANODE / Type: RIGAKU / Wavelength: 1.5418 |
| Detector | Type: XUONG-HAMLIN MULTIWIRE / Detector: AREA DETECTOR / Date: May 1, 1997 |
| Radiation | Monochromator: QUARTZ CRYSTAL / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2.36→50 Å / Num. obs: 22616 / % possible obs: 91.6 % / Observed criterion σ(I): 2 / Redundancy: 3.6 % / Rsym value: 0.098 / Net I/σ(I): 9.7 |
| Reflection shell | Resolution: 2.36→2.55 Å / Redundancy: 2.3 % / Mean I/σ(I) obs: 2.8 / Rsym value: 0.277 / % possible all: 76.1 |
| Reflection | *PLUS Num. measured all: 81247 / Rmerge(I) obs: 0.098 |
| Reflection shell | *PLUS % possible obs: 76.1 % / Num. unique obs: 3671 / Num. measured obs: 8466 / Rmerge(I) obs: 0.277 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1IGM Resolution: 2.36→6 Å / Rfactor Rfree error: 0.0073 / Data cutoff high absF: 10000000 / Data cutoff low absF: 3 / Isotropic thermal model: RESTRAINED (GAUSSIAN) / Cross valid method: THROUGHOUT / σ(F): 2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 10.77 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.36→6 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.36→2.46 Å / Rfactor Rfree error: 0.039 / Total num. of bins used: 8
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Version: 3.851 / Classification: refinement X-PLOR / Version: 3.851 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor obs: 0.206 / Rfactor Rfree: 0.285 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Rfactor Rfree: 0.373 / Rfactor obs: 0.2582 |
 Movie
Movie Controller
Controller












 PDBj
PDBj



