[English] 日本語
 Yorodumi
Yorodumi- PDB-4ac5: Lipidic sponge phase crystal structure of the Bl. viridis reactio... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4ac5 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Lipidic sponge phase crystal structure of the Bl. viridis reaction centre solved using serial femtosecond crystallography | ||||||
 Components Components |
| ||||||
 Keywords Keywords | PHOTOSYNTHESIS / LIPIDIC-SPONGE PHASE / REACTION CENTER / ELECTRON TRANSPORT / CELL MEMBRANE / METAL- BINDING / TRANSMEMBRANE / FORMYLATION / CHROMOPHORE / LIPOPROTEIN | ||||||
| Function / homology |  Function and homology information Function and homology informationplasma membrane-derived chromatophore membrane / plasma membrane light-harvesting complex / bacteriochlorophyll binding / photosynthetic electron transport in photosystem II / : / photosynthesis, light reaction / photosynthesis / electron transfer activity / iron ion binding / heme binding / metal ion binding Similarity search - Function | ||||||
| Biological species |  BLASTOCHLORIS VIRIDIS (bacteria) BLASTOCHLORIS VIRIDIS (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  FREE ELECTRON LASER / FREE ELECTRON LASER /  MOLECULAR REPLACEMENT / Resolution: 8.2 Å MOLECULAR REPLACEMENT / Resolution: 8.2 Å | ||||||
 Authors Authors | Johansson, L.C. / Arnlund, D. / White, T.A. / Katona, G. / DePonte, D.P. / Weierstall, U. / Doak, R.B. / Shoeman, R.L. / Lomb, L. / Malmerberg, E. ...Johansson, L.C. / Arnlund, D. / White, T.A. / Katona, G. / DePonte, D.P. / Weierstall, U. / Doak, R.B. / Shoeman, R.L. / Lomb, L. / Malmerberg, E. / Davidsson, J. / Nass, K. / Liang, M. / Andreasson, J. / Aquila, A. / Bajt, S. / Barthelmess, M. / Barty, A. / Bogan, M.J. / Bostedt, C. / Bozek, J.D. / Caleman, C. / Coffee, R. / Coppola, N. / Ekeberg, T. / Epp, S.W. / Erk, B. / Fleckenstein, H. / Foucar, L. / Graafsma, H. / Gumprecht, L. / Hajdu, J. / Hampton, C.Y. / Hartmann, R. / Hartmann, A. / Hauser, G. / Hirsemann, H. / Holl, P. / Hunter, M.S. / Kassemeyer, S. / Kimmel, N. / Kirian, R.A. / Maia, F.R.N.C. / Marchesini, S. / Martin, A.V. / Reich, C. / Rolles, D. / Rudek, B. / Rudenko, A. / Schlichting, I. / Schulz, J. / Seibert, M.M. / Sierra, R. / Soltau, H. / Starodub, D. / Stellato, F. / Stern, S. / Struder, L. / Timneanu, N. / Ullrich, J. / Wahlgren, W.Y. / Wang, X. / Weidenspointner, G. / Wunderer, C. / Fromme, P. / Chapman, H.N. / Spence, J.C.H. / Neutze, R. | ||||||
 Citation Citation |  Journal: Nat.Methods / Year: 2012 Journal: Nat.Methods / Year: 2012Title: Lipidic Phase Membrane Protein Serial Femtosecond Crystallography. Authors: Johansson, L.C. / Arnlund, D. / White, T.A. / Katona, G. / Deponte, D.P. / Weierstall, U. / Doak, R.B. / Shoeman, R.L. / Lomb, L. / Malmerberg, E. / Davidsson, J. / Nass, K. / Liang, M. / ...Authors: Johansson, L.C. / Arnlund, D. / White, T.A. / Katona, G. / Deponte, D.P. / Weierstall, U. / Doak, R.B. / Shoeman, R.L. / Lomb, L. / Malmerberg, E. / Davidsson, J. / Nass, K. / Liang, M. / Andreasson, J. / Aquila, A. / Bajt, S. / Barthelmess, M. / Barty, A. / Bogan, M.J. / Bostedt, C. / Bozek, J.D. / Caleman, C. / Coffee, R. / Coppola, N. / Ekeberg, T. / Epp, S.W. / Erk, B. / Fleckenstein, H. / Foucar, L. / Graafsma, H. / Gumprecht, L. / Hajdu, J. / Hampton, C.Y. / Hartmann, R. / Hartmann, A. / Hauser, G. / Hirsemann, H. / Holl, P. / Hunter, M.S. / Kassemeyer, S. / Kimmel, N. / Kirian, R.A. / Maia, F.R.N.C. / Marchesini, S. / Martin, A.V. / Reich, C. / Rolles, D. / Rudek, B. / Rudenko, A. / Schlichting, I. / Schulz, J. / Seibert, M.M. / Sierra, R. / Soltau, H. / Starodub, D. / Stellato, F. / Stern, S. / Struder, L. / Timneanu, N. / Ullrich, J. / Wahlgren, W.Y. / Wang, X. / Weidenspointner, G. / Wunderer, C. / Fromme, P. / Chapman, H.N. / Spence, J.C.H. / Neutze, R. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4ac5.cif.gz 4ac5.cif.gz | 254.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4ac5.ent.gz pdb4ac5.ent.gz | 199.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4ac5.json.gz 4ac5.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  4ac5_validation.pdf.gz 4ac5_validation.pdf.gz | 2 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  4ac5_full_validation.pdf.gz 4ac5_full_validation.pdf.gz | 2 MB | Display | |
| Data in XML |  4ac5_validation.xml.gz 4ac5_validation.xml.gz | 48.6 KB | Display | |
| Data in CIF |  4ac5_validation.cif.gz 4ac5_validation.cif.gz | 62.9 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ac/4ac5 https://data.pdbj.org/pub/pdb/validation_reports/ac/4ac5 ftp://data.pdbj.org/pub/pdb/validation_reports/ac/4ac5 ftp://data.pdbj.org/pub/pdb/validation_reports/ac/4ac5 | HTTPS FTP |
-Related structure data
| Related structure data |  2wjnS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 1 molecules C
| #1: Protein | Mass: 37450.801 Da / Num. of mol.: 1 / Source method: isolated from a natural source Details: DEUTSCHE SAMMLUNG VON MIKROORGANISMEN UND ZELLKULTUREN GMBH (DSMZ) Source: (natural)  BLASTOCHLORIS VIRIDIS (bacteria) / References: UniProt: P07173 BLASTOCHLORIS VIRIDIS (bacteria) / References: UniProt: P07173 |
|---|
-REACTION CENTER PROTEIN ... , 3 types, 3 molecules HLM
| #2: Protein | Mass: 28557.453 Da / Num. of mol.: 1 / Source method: isolated from a natural source Details: DEUTSCHE SAMMLUNG VON MIKROORGANISMEN UND ZELLKULTUREN GMBH (DSMZ) Source: (natural)  BLASTOCHLORIS VIRIDIS (bacteria) / References: UniProt: P06008 BLASTOCHLORIS VIRIDIS (bacteria) / References: UniProt: P06008 |
|---|---|
| #3: Protein | Mass: 30600.299 Da / Num. of mol.: 1 / Source method: isolated from a natural source Details: DEUTSCHE SAMMLUNG VON MIKROORGANISMEN UND ZELLKULTUREN GMBH (DSMZ) Source: (natural)  BLASTOCHLORIS VIRIDIS (bacteria) / References: UniProt: P06009 BLASTOCHLORIS VIRIDIS (bacteria) / References: UniProt: P06009 |
| #4: Protein | Mass: 36091.395 Da / Num. of mol.: 1 / Source method: isolated from a natural source Details: DEUTSCHE SAMMLUNG VON MIKROORGANISMEN UND ZELLKULTUREN GMBH (DSMZ) Source: (natural)  BLASTOCHLORIS VIRIDIS (bacteria) / References: UniProt: P06010 BLASTOCHLORIS VIRIDIS (bacteria) / References: UniProt: P06010 |
-Non-polymers , 6 types, 13 molecules 
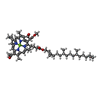









| #5: Chemical | ChemComp-HEM / #6: Chemical | ChemComp-BCB / #7: Chemical | #8: Chemical | ChemComp-FE2 / | #9: Chemical | ChemComp-MQ7 / | #10: Chemical | ChemComp-NS5 / | |
|---|
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 265 X-RAY DIFFRACTION / Number of used crystals: 265 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.4 Å3/Da / Density % sol: 64 % Description: THE DATA WERE COLLECTED AT THE LINAC COHERENT LIGHT SOURCE USING SERIAL FEMTOSECOND CRYSTALLOGRAPHY TO PROVE THAT THE SPONGE PHASE CRYSTALLISATION METHOD IS COMPATABLE WITH SERIAL ...Description: THE DATA WERE COLLECTED AT THE LINAC COHERENT LIGHT SOURCE USING SERIAL FEMTOSECOND CRYSTALLOGRAPHY TO PROVE THAT THE SPONGE PHASE CRYSTALLISATION METHOD IS COMPATABLE WITH SERIAL FEMTOSECOND CRYSTALLOGRAPHY. THE DATA WAS MERGED FROM 265 MICROCRYSTALS USING THE IN-HOUSE PROGRAM CRYSTFEL 0.1.0 AND INDEXAMAJIG. |
|---|---|
| Crystal grow | pH: 7.9 Details: BATCH CRYSTALLIZATIONS WERE SET UP IN SEPTUM-SEALED GLASS VIALS CONTAINING 100 UL PROTEIN (20-30 MG/ML) , 100 UL LIPIDIC SPONGE PHASE (12 % MONOOLEIN, 17.5 % JEFFAMINE M- 600, 1.0 M HEPES PH ...Details: BATCH CRYSTALLIZATIONS WERE SET UP IN SEPTUM-SEALED GLASS VIALS CONTAINING 100 UL PROTEIN (20-30 MG/ML) , 100 UL LIPIDIC SPONGE PHASE (12 % MONOOLEIN, 17.5 % JEFFAMINE M- 600, 1.0 M HEPES PH 8.0, 0.7 M (NH4)2SO4, 2.5 % 1,2,3- HEPTANETRIOL) AND 50 UL 1.0-1.2 M TRI-SODIUM CITRATE |
-Data collection
| Diffraction | Mean temperature: 288 K / Serial crystal experiment: Y |
|---|---|
| Diffraction source | Source:  FREE ELECTRON LASER / Site: FREE ELECTRON LASER / Site:  SLAC LCLS SLAC LCLS  / Beamline: AMO / Wavelength: 6.2 / Wavelength: 6.2 Å / Beamline: AMO / Wavelength: 6.2 / Wavelength: 6.2 Å |
| Detector | Type: ADSC QUANTUM 315r / Detector: CCD / Date: Jun 14, 2010 |
| Radiation | Monochromator: SI 111 / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 6.2 Å / Relative weight: 1 |
| Reflection | Resolution: 7.4→46.1 Å / Num. obs: 2431 / % possible obs: 85 % / Observed criterion σ(I): 2 / Redundancy: 8 % / Rmerge(I) obs: 0.5 / Net I/σ(I): 1.9 |
| Reflection shell | Resolution: 7.37→7.62 Å / Redundancy: 1.1 % / % possible all: 21.1 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 2WJN Resolution: 8.2→46.1 Å / Cor.coef. Fo:Fc: 0.665 / SU ML: 9.332 / Cross valid method: THROUGHOUT / ESU R Free: 7.361 / Stereochemistry target values: MAXIMUM LIKELIHOOD Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS. U VALUES REFINED INDIVIDUALLY.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 15.401 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 8.2→46.1 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller























 PDBj
PDBj















