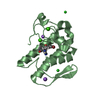
Entry Database : PDB / ID : 3zy0Title Crystal structure of a truncated variant of the human p63 tetramerization domain lacking the C-terminal helix TUMOR PROTEIN P63 Keywords / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species HOMO SAPIENS (human)Method / / / Resolution : 1.9 Å Authors Joerger, A.C. Journal : J.Mol.Biol. / Year : 2012Title : Structure and Kinetic Stability of the P63 Tetramerization Domain.Authors : Natan, E. / Joerger, A.C. History Deposition Aug 16, 2011 Deposition site / Processing site Revision 1.0 Nov 30, 2011 Provider / Type Revision 1.1 Jan 25, 2012 Group Revision 1.2 May 8, 2019 Group Data collection / Derived calculations ... Data collection / Derived calculations / Experimental preparation / Other Category database_PDB_rev / database_PDB_rev_record ... database_PDB_rev / database_PDB_rev_record / exptl_crystal_grow / pdbx_database_proc / pdbx_database_status / struct_conn Item _exptl_crystal_grow.method / _exptl_crystal_grow.temp ... _exptl_crystal_grow.method / _exptl_crystal_grow.temp / _pdbx_database_status.recvd_author_approval / _struct_conn.pdbx_leaving_atom_flag Revision 1.3 Oct 23, 2024 Group / Database references / Structure summaryCategory chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_entry_details / pdbx_modification_feature Item / _database_2.pdbx_database_accession
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information HOMO SAPIENS (human)
HOMO SAPIENS (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MAD / Resolution: 1.9 Å
MAD / Resolution: 1.9 Å  Authors
Authors Citation
Citation Journal: J.Mol.Biol. / Year: 2012
Journal: J.Mol.Biol. / Year: 2012 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 3zy0.cif.gz
3zy0.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb3zy0.ent.gz
pdb3zy0.ent.gz PDB format
PDB format 3zy0.json.gz
3zy0.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/zy/3zy0
https://data.pdbj.org/pub/pdb/validation_reports/zy/3zy0 ftp://data.pdbj.org/pub/pdb/validation_reports/zy/3zy0
ftp://data.pdbj.org/pub/pdb/validation_reports/zy/3zy0 Links
Links Assembly
Assembly
 Components
Components HOMO SAPIENS (human) / Production host:
HOMO SAPIENS (human) / Production host: 
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  Diamond
Diamond  / Beamline: I02 / Wavelength: 0.9796
/ Beamline: I02 / Wavelength: 0.9796  Processing
Processing MAD
MAD Movie
Movie Controller
Controller















 PDBj
PDBj








