[English] 日本語
 Yorodumi
Yorodumi- PDB-3zfs: Cryo-EM structure of the F420-reducing NiFe-hydrogenase from a me... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3zfs | ||||||
|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the F420-reducing NiFe-hydrogenase from a methanogenic archaeon with bound substrate | ||||||
 Components Components | (F420-REDUCING HYDROGENASE, SUBUNIT ...) x 3 | ||||||
 Keywords Keywords | OXIDOREDUCTASE / METHANOGENESIS | ||||||
| Function / homology |  Function and homology information Function and homology informationcoenzyme F420 hydrogenase / coenzyme F420 hydrogenase activity / oxidoreductase activity, acting on CH or CH2 groups, with an iron-sulfur protein as acceptor / ferredoxin hydrogenase activity / nickel cation binding / iron-sulfur cluster binding / flavin adenine dinucleotide binding / 4 iron, 4 sulfur cluster binding Similarity search - Function | ||||||
| Biological species |   METHANOTHERMOBACTER MARBURGENSIS (archaea) METHANOTHERMOBACTER MARBURGENSIS (archaea) | ||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 4 Å | ||||||
| Model type details | CA ATOMS ONLY, CHAIN A, B, C | ||||||
 Authors Authors | Mills, D.J. / Vitt, S. / Strauss, M. / Shima, S. / Vonck, J. | ||||||
 Citation Citation |  Journal: Elife / Year: 2013 Journal: Elife / Year: 2013Title: De novo modeling of the F(420)-reducing [NiFe]-hydrogenase from a methanogenic archaeon by cryo-electron microscopy. Authors: Deryck J Mills / Stella Vitt / Mike Strauss / Seigo Shima / Janet Vonck /  Abstract: Methanogenic archaea use a [NiFe]-hydrogenase, Frh, for oxidation/reduction of F420, an important hydride carrier in the methanogenesis pathway from H2 and CO2. Frh accounts for about 1% of the ...Methanogenic archaea use a [NiFe]-hydrogenase, Frh, for oxidation/reduction of F420, an important hydride carrier in the methanogenesis pathway from H2 and CO2. Frh accounts for about 1% of the cytoplasmic protein and forms a huge complex consisting of FrhABG heterotrimers with each a [NiFe] center, four Fe-S clusters and an FAD. Here, we report the structure determined by near-atomic resolution cryo-EM of Frh with and without bound substrate F420. The polypeptide chains of FrhB, for which there was no homolog, was traced de novo from the EM map. The 1.2-MDa complex contains 12 copies of the heterotrimer, which unexpectedly form a spherical protein shell with a hollow core. The cryo-EM map reveals strong electron density of the chains of metal clusters running parallel to the protein shell, and the F420-binding site is located at the end of the chain near the outside of the spherical structure. DOI:http://dx.doi.org/10.7554/eLife.00218.001. | ||||||
| History |
| ||||||
| Remark 650 | HELIX DETERMINATION METHOD: AUTHOR PROVIDED. | ||||||
| Remark 700 | SHEET DETERMINATION METHOD: AUTHOR PROVIDED. |
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3zfs.cif.gz 3zfs.cif.gz | 52.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3zfs.ent.gz pdb3zfs.ent.gz | 26.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3zfs.json.gz 3zfs.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  3zfs_validation.pdf.gz 3zfs_validation.pdf.gz | 1.2 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  3zfs_full_validation.pdf.gz 3zfs_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  3zfs_validation.xml.gz 3zfs_validation.xml.gz | 21.7 KB | Display | |
| Data in CIF |  3zfs_validation.cif.gz 3zfs_validation.cif.gz | 29.9 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/zf/3zfs https://data.pdbj.org/pub/pdb/validation_reports/zf/3zfs ftp://data.pdbj.org/pub/pdb/validation_reports/zf/3zfs ftp://data.pdbj.org/pub/pdb/validation_reports/zf/3zfs | HTTPS FTP |
-Related structure data
| Related structure data |  2096MC  2097MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-F420-REDUCING HYDROGENASE, SUBUNIT ... , 3 types, 3 molecules ABC
| #1: Protein | Mass: 44873.332 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Details: FRHA CONTAINS A NIFE CENTER Source: (natural)   METHANOTHERMOBACTER MARBURGENSIS (archaea) METHANOTHERMOBACTER MARBURGENSIS (archaea)Strain: DSM 2133 / References: UniProt: D9PYF9, coenzyme F420 hydrogenase |
|---|---|
| #2: Protein | Mass: 30267.762 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Details: FRHG CONTAINS THREE 4FE4S CLUSTERS Source: (natural)   METHANOTHERMOBACTER MARBURGENSIS (archaea) METHANOTHERMOBACTER MARBURGENSIS (archaea)Strain: DSM 2133 / References: UniProt: D9PYF7, coenzyme F420 hydrogenase |
| #3: Protein | Mass: 30778.752 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Details: FRHB CONTAINS FAD AND A 4FE4S CLUSTER Source: (natural)   METHANOTHERMOBACTER MARBURGENSIS (archaea) METHANOTHERMOBACTER MARBURGENSIS (archaea)Strain: DSM 2133 / References: UniProt: D9PYF6, coenzyme F420 hydrogenase |
-Non-polymers , 6 types, 9 molecules 



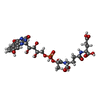






| #4: Chemical | ChemComp-FCO / | ||||
|---|---|---|---|---|---|
| #5: Chemical | ChemComp-NI / | ||||
| #6: Chemical | ChemComp-FE2 / | ||||
| #7: Chemical | ChemComp-SF4 / #8: Chemical | ChemComp-F42 / | #9: Chemical | ChemComp-FAD / | |
-Details
| Nonpolymer details | COENZYME F420 (F42): ONLY ISOALLOXAZ |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: F420-REDUCING HYDROGENASE / Type: COMPLEX |
|---|---|
| Buffer solution | Name: 50 MM TRIS/HCL, 2 MM DTT, 0.025 MM FAD / pH: 7.6 / Details: 50 MM TRIS/HCL, 2 MM DTT, 0.025 MM FAD |
| Specimen | Conc.: 0.7 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Specimen support | Details: HOLEY CARBON |
| Vitrification | Instrument: FEI VITROBOT MARK I / Cryogen name: ETHANE / Details: LIQUID ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI POLARA 300 / Date: Feb 11, 2011 |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 200 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 200 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 59000 X / Calibrated magnification: 61400 X / Nominal defocus max: 3820 nm / Nominal defocus min: 1650 nm / Cs: 2 mm |
| Specimen holder | Temperature: 77 K |
| Image recording | Electron dose: 15 e/Å2 / Film or detector model: KODAK SO-163 FILM |
| Image scans | Num. digital images: 80 |
| Radiation wavelength | Relative weight: 1 |
- Processing
Processing
| EM software |
| ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Details: PER MICROGRAPH | ||||||||||||||||
| Symmetry | Point symmetry: T (tetrahedral) | ||||||||||||||||
| 3D reconstruction | Method: REFINEMENT IN EMAN2 / Resolution: 4 Å / Num. of particles: 97290 / Nominal pixel size: 1.19 Å / Actual pixel size: 1.14 Å / Magnification calibration: FIT OF X-RAY MODEL Details: SUBMISSION BASED ON EXPERIMENTAL DATA FROM EMDB EMD-2097.(DEPOSITION ID: 10788). Symmetry type: POINT | ||||||||||||||||
| Atomic model building | Protocol: RIGID BODY FIT / Space: REAL / Details: REFINEMENT PROTOCOL--RIGID BODY | ||||||||||||||||
| Atomic model building | PDB-ID: 2WPN Accession code: 2WPN / Source name: PDB / Type: experimental model | ||||||||||||||||
| Refinement | Highest resolution: 4 Å | ||||||||||||||||
| Refinement step | Cycle: LAST / Highest resolution: 4 Å
|
 Movie
Movie Controller
Controller












 PDBj
PDBj

















