+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3s5w | ||||||
|---|---|---|---|---|---|---|---|
| Title | Ornithine Hydroxylase (PvdA) from Pseudomonas aeruginosa | ||||||
 Components Components | L-ornithine 5-monooxygenase | ||||||
 Keywords Keywords | OXIDOREDUCTASE / CLASS B FLAVIN DEPENDENT N-HYDROXYLATING MONOOXYGENASE / Class B Flavin Dependent Monooxygenase N-Hydroxylating / Monooxygenase / Bacterial cytosol | ||||||
| Function / homology |  Function and homology information Function and homology informationL-ornithine N5-monooxygenase (NADPH) / ornithine N5-monooxygenase activity / pyoverdine biosynthetic process / intracellular iron ion homeostasis / plasma membrane / cytoplasm Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.9 Å MOLECULAR REPLACEMENT / Resolution: 1.9 Å | ||||||
 Authors Authors | Olucha, J. / Lamb, A.L. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2011 Journal: J.Biol.Chem. / Year: 2011Title: Two Structures of an N-Hydroxylating Flavoprotein Monooxygenase: ORNITHINE HYDROXYLASE FROM PSEUDOMONAS AERUGINOSA. Authors: Olucha, J. / Meneely, K.M. / Chilton, A.S. / Lamb, A.L. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3s5w.cif.gz 3s5w.cif.gz | 190.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3s5w.ent.gz pdb3s5w.ent.gz | 149.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3s5w.json.gz 3s5w.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/s5/3s5w https://data.pdbj.org/pub/pdb/validation_reports/s5/3s5w ftp://data.pdbj.org/pub/pdb/validation_reports/s5/3s5w ftp://data.pdbj.org/pub/pdb/validation_reports/s5/3s5w | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
| ||||||||
| Details | AUTHORS STATE THAT DYNAMIC LIGHT SCATTERING EXPERIMENTS HAVE SHOWN PVDA BEHAVES AS A MONOMER IN SOLUTION (MENEELY, K.M. AND LAMB, A.L., BIOCHEMISTRY, 2007, 46 (42), 11930-11937). |
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 51711.164 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   References: UniProt: Q51548, Oxidoreductases; Acting on single donors with incorporation of molecular oxygen (oxygenases); With incorporation of one atom of oxygen (internal monooxygenases or ...References: UniProt: Q51548, Oxidoreductases; Acting on single donors with incorporation of molecular oxygen (oxygenases); With incorporation of one atom of oxygen (internal monooxygenases or internal mixed-function oxidases) |
|---|
-Non-polymers , 5 types, 385 molecules 

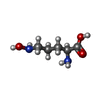






| #2: Chemical | | #3: Chemical | #4: Chemical | #5: Chemical | #6: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.3 Å3/Da / Density % sol: 62.73 % |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, hanging drop / pH: 5 Details: 9.5% PEG 8000, 25% glycerol, 60 mM potassium phosphate monobasic, pH 5.0, VAPOR DIFFUSION, HANGING DROP, temperature 291K |
-Data collection
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRL SSRL  / Beamline: BL9-2 / Wavelength: 1 Å / Beamline: BL9-2 / Wavelength: 1 Å |
|---|---|
| Detector | Type: MARMOSAIC 325 mm CCD / Detector: CCD / Date: Jul 17, 2009 |
| Radiation | Monochromator: Double Crystal Monochromator / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 1.9→36.8 Å / Num. all: 111425 / Num. obs: 108180 / % possible obs: 99.7 % |
| Reflection shell | Resolution: 1.9→2 Å / Rmerge(I) obs: 0.412 / Mean I/σ(I) obs: 4.1 / % possible all: 97.5 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 2.8 A Seleno-SAD PVDA MODEL Resolution: 1.9→36.74 Å / Cor.coef. Fo:Fc: 0.954 / Cor.coef. Fo:Fc free: 0.944 / SU B: 2.441 / SU ML: 0.073 / Cross valid method: THROUGHOUT / ESU R Free: 0.111 Stereochemistry target values: MAXIMUM LIKELIHOOD WITH PHASES Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: BABINET MODEL WITH MASK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 31.838 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.9→36.74 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.9→1.947 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller














 PDBj
PDBj



