[English] 日本語
 Yorodumi
Yorodumi- PDB-3r1a: Closed crystal structure of cytochrome P450 2B4 covalently bound ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3r1a | ||||||
|---|---|---|---|---|---|---|---|
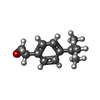
| Title | Closed crystal structure of cytochrome P450 2B4 covalently bound to the mechanism-based inactivator tert-butylphenylacetylene | ||||||
 Components Components | Cytochrome P450 2B4 | ||||||
 Keywords Keywords | OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR / cytochrome P450 2B4 / monooxygenase / oxidoreductase / membrane protein / T302 covalently linked to tert-butylphenylacetylene / OXIDOREDUCTASE-OXIDOREDUCTASE INHIBITOR complex | ||||||
| Function / homology |  Function and homology information Function and homology informationarachidonate epoxygenase activity / epoxygenase P450 pathway / oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen / unspecific monooxygenase / xenobiotic metabolic process / iron ion binding / heme binding / endoplasmic reticulum membrane Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 3.5 Å molecular replacement / Resolution: 3.5 Å | ||||||
 Authors Authors | Gay, S.C. / Zhang, H. / Stout, C.D. / Hollenberg, P.F. / Halpert, J.R. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 2011 Journal: Biochemistry / Year: 2011Title: Structural Analysis of Mammalian Cytochrome P450 2B4 Covalently Bound to the Mechanism-Based Inactivator tert-Butylphenylacetylene: Insight into Partial Enzymatic Activity. Authors: Gay, S.C. / Zhang, H. / Wilderman, P.R. / Roberts, A.G. / Liu, T. / Li, S. / Lin, H.L. / Zhang, Q. / Woods, V.L. / Stout, C.D. / Hollenberg, P.F. / Halpert, J.R. #1:  Journal: J.Biol.Chem. / Year: 2006 Journal: J.Biol.Chem. / Year: 2006Title: Structure of microsomal cytochrome P450 2B4 complexed with the antifungal drug bifonazole: insight into P450 conformational plasticity and membrane interaction. Authors: Zhao, Y. / White, M.A. / Muralidhara, B.K. / Sun, L. / Halpert, J.R. / Stout, C.D. #2:  Journal: J.Biol.Chem. / Year: 2004 Journal: J.Biol.Chem. / Year: 2004Title: Structure of mammalian cytochrome P450 2B4 complexed with 4-(4-chlorophenyl)imidazole at 1.9-A resolution: insight into the range of P450 conformations and the coordination of redox partner binding. Authors: Scott, E.E. / White, M.A. / He, Y.A. / Johnson, E.F. / Stout, C.D. / Halpert, J.R. #3:  Journal: Proc.Natl.Acad.Sci.USA / Year: 2003 Journal: Proc.Natl.Acad.Sci.USA / Year: 2003Title: An open conformation of mammalian cytochrome P450 2B4 at 1.6-A resolution. Authors: Scott, E.E. / He, Y.A. / Wester, M.R. / White, M.A. / Chin, C.C. / Halpert, J.R. / Johnson, E.F. / Stout, C.D. #4:  Journal: Biochemistry / Year: 2007 Journal: Biochemistry / Year: 2007Title: Structural and thermodynamic consequences of 1-(4-chlorophenyl)imidazole binding to cytochrome P450 2B4. Authors: Zhao, Y. / Sun, L. / Muralidhara, B.K. / Kumar, S. / White, M.A. / Stout, C.D. / Halpert, J.R. #5:  Journal: Biochemistry / Year: 2009 Journal: Biochemistry / Year: 2009Title: Crystal structures of cytochrome P450 2B4 in complex with the inhibitor 1-biphenyl-4-methyl-1H-imidazole: ligand-induced structural response through alpha-helical repositioning. Authors: Gay, S.C. / Sun, L. / Maekawa, K. / Halpert, J.R. / Stout, C.D. #6:  Journal: Biochemistry / Year: 2010 Journal: Biochemistry / Year: 2010Title: Structures of cytochrome P450 2B4 complexed with the antiplatelet drugs ticlopidine and clopidogrel . Authors: Gay, S.C. / Roberts, A.G. / Maekawa, K. / Talakad, J.C. / Hong, W.X. / Zhang, Q. / Stout, C.D. / Halpert, J.R. #7:  Journal: J.Biol.Chem. / Year: 2010 Journal: J.Biol.Chem. / Year: 2010Title: Plasticity of cytochrome P450 2B4 as investigated by hydrogen-deuterium exchange mass spectrometry and X-ray crystallography. Authors: Wilderman, P.R. / Shah, M.B. / Liu, T. / Li, S. / Hsu, S. / Roberts, A.G. / Goodlett, D.R. / Zhang, Q. / Woods, V.L. / Stout, C.D. / Halpert, J.R. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3r1a.cif.gz 3r1a.cif.gz | 714.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3r1a.ent.gz pdb3r1a.ent.gz | 583.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3r1a.json.gz 3r1a.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/r1/3r1a https://data.pdbj.org/pub/pdb/validation_reports/r1/3r1a ftp://data.pdbj.org/pub/pdb/validation_reports/r1/3r1a ftp://data.pdbj.org/pub/pdb/validation_reports/r1/3r1a | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3r1bC  1suoS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| 4 | 
| ||||||||
| 5 | 
| ||||||||
| 6 | 
| ||||||||
| 7 | 
| ||||||||
| 8 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 54169.082 Da / Num. of mol.: 8 Mutation: E2A, G22K, H23K, P24T, K25S, A26S, H27K, R29K, P221S, H226Y Source method: isolated from a genetically manipulated source Source: (gene. exp.)   #2: Chemical | ChemComp-HEM / #3: Chemical | ChemComp-TB2 / ( #4: Water | ChemComp-HOH / | Has protein modification | Y | Nonpolymer details | THE PROTEIN WAS INCUBATED WITH TERT-BUTYLPHENYLACETYLENE, CATALASE, CYTOCHROME P450 REDUCTASE, AND ...THE PROTEIN WAS INCUBATED WITH TERT-BUTYLPHENY | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.57 Å3/Da / Density % sol: 52.13 % |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, sitting drop / pH: 7.4 Details: 0.1 M magnesium formate, 15% (w/v) PEG 3350, pH 7.4, vapor diffusion, sitting drop, temperature 291K |
-Data collection
| Diffraction | Mean temperature: 100 K | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRL SSRL  / Beamline: BL11-1 / Wavelength: 0.98 Å / Beamline: BL11-1 / Wavelength: 0.98 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: MARMOSAIC 325 mm CCD / Detector: CCD / Date: Jan 22, 2010 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Monochromator: Si(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 0.98 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 3.5→229.275 Å / Num. all: 51302 / Num. obs: 51302 / % possible obs: 90.2 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 3.2 % / Biso Wilson estimate: 72.64 Å2 / Rmerge(I) obs: 0.156 / Rsym value: 0.156 / Net I/σ(I): 6.5 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell |
|
-Phasing
| Phasing | Method:  molecular replacement molecular replacement | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Phasing MR | Rfactor: 44.43 / Model details: Phaser MODE: MR_AUTO
|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1SUO Resolution: 3.5→115.93 Å / Occupancy max: 1 / Occupancy min: 1 / SU ML: 0.46 / σ(F): 0 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.27 Å / VDW probe radii: 0.6 Å / Solvent model: FLAT BULK SOLVENT MODEL / Bsol: 53.456 Å2 / ksol: 0.349 e/Å3 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 159.12 Å2 / Biso mean: 73.8183 Å2 / Biso min: 29.65 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3.5→115.93 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 18
|
 Movie
Movie Controller
Controller






















 PDBj
PDBj