[English] 日本語
 Yorodumi
Yorodumi- PDB-3hs7: X-ray crystal structure of docosahexaenoic acid bound to the cycl... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3hs7 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | X-ray crystal structure of docosahexaenoic acid bound to the cyclooxygenase channel of cyclooxygenase-2 | |||||||||
 Components Components | Prostaglandin G/H synthase 2 | |||||||||
 Keywords Keywords | OXIDOREDUCTASE / Dioxygenase / Disulfide bond / Endoplasmic reticulum / Fatty acid biosynthesis / Glycoprotein / Heme / Iron / Lipid synthesis / Membrane / Metal-binding / Microsome / Peroxidase / Phosphoprotein / Prostaglandin biosynthesis | |||||||||
| Function / homology |  Function and homology information Function and homology informationBiosynthesis of DHA-derived SPMs / Biosynthesis of EPA-derived SPMs / Biosynthesis of DPAn-3 SPMs / Biosynthesis of electrophilic ω-3 PUFA oxo-derivatives / Synthesis of 15-eicosatetraenoic acid derivatives / cellular response to non-ionic osmotic stress / : / Synthesis of Prostaglandins (PG) and Thromboxanes (TX) / prostaglandin-endoperoxide synthase / prostaglandin-endoperoxide synthase activity ...Biosynthesis of DHA-derived SPMs / Biosynthesis of EPA-derived SPMs / Biosynthesis of DPAn-3 SPMs / Biosynthesis of electrophilic ω-3 PUFA oxo-derivatives / Synthesis of 15-eicosatetraenoic acid derivatives / cellular response to non-ionic osmotic stress / : / Synthesis of Prostaglandins (PG) and Thromboxanes (TX) / prostaglandin-endoperoxide synthase / prostaglandin-endoperoxide synthase activity / cyclooxygenase pathway / negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress / positive regulation of prostaglandin biosynthetic process / regulation of neuroinflammatory response / positive regulation of fever generation / response to selenium ion / prostaglandin secretion / cellular response to fluid shear stress / response to nematode / prostaglandin biosynthetic process / nuclear inner membrane / dioxygenase activity / bone mineralization / decidualization / nuclear outer membrane / keratinocyte differentiation / positive regulation of brown fat cell differentiation / brown fat cell differentiation / embryo implantation / peroxidase activity / regulation of blood pressure / regulation of cell population proliferation / response to oxidative stress / cellular response to hypoxia / neuron projection / heme binding / endoplasmic reticulum membrane / negative regulation of apoptotic process / enzyme binding / protein homodimerization activity / metal ion binding / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.65 Å MOLECULAR REPLACEMENT / Resolution: 2.65 Å | |||||||||
 Authors Authors | Vecchio, A.J. / Simmons, D.M. / Malkowski, M.G. | |||||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2010 Journal: J.Biol.Chem. / Year: 2010Title: Structural basis of fatty acid substrate binding to cyclooxygenase-2. Authors: Vecchio, A.J. / Simmons, D.M. / Malkowski, M.G. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3hs7.cif.gz 3hs7.cif.gz | 401.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3hs7.ent.gz pdb3hs7.ent.gz | 318.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3hs7.json.gz 3hs7.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/hs/3hs7 https://data.pdbj.org/pub/pdb/validation_reports/hs/3hs7 ftp://data.pdbj.org/pub/pdb/validation_reports/hs/3hs7 ftp://data.pdbj.org/pub/pdb/validation_reports/hs/3hs7 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3hs5C  3hs6C  3krkC  1cvuS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 67933.391 Da / Num. of mol.: 2 / Mutation: N580A Source method: isolated from a genetically manipulated source Source: (gene. exp.)   References: UniProt: Q05769, prostaglandin-endoperoxide synthase |
|---|
-Sugars , 4 types, 8 molecules 


| #2: Polysaccharide | Source method: isolated from a genetically manipulated source #3: Polysaccharide | alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta- ...alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | Source method: isolated from a genetically manipulated source #6: Sugar | #7: Sugar | |
|---|
-Non-polymers , 5 types, 345 molecules 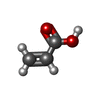
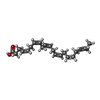






| #4: Chemical | | #5: Chemical | #8: Chemical | #9: Chemical | ChemComp-EDO / #10: Water | ChemComp-HOH / | |
|---|
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.59 Å3/Da / Density % sol: 52.53 % |
|---|---|
| Crystal grow | Temperature: 296 K / Method: vapor diffusion, sitting drop / pH: 7.5 Details: 23-34% Polyacrylic acid 5100, 100mM HEPES pH 7.5, 20mM MgCl2, VAPOR DIFFUSION, SITTING DROP, temperature 296K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  CHESS CHESS  / Beamline: A1 / Wavelength: 0.9789 Å / Beamline: A1 / Wavelength: 0.9789 Å |
| Detector | Type: ADSC QUANTUM 210 / Detector: CCD / Date: Dec 21, 2008 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9789 Å / Relative weight: 1 |
| Reflection | Resolution: 2.65→20 Å / Num. obs: 41205 / % possible obs: 99.8 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 5.9 % / Biso Wilson estimate: 50.4 Å2 / Rmerge(I) obs: 0.124 / Net I/σ(I): 10.9 |
| Reflection shell | Resolution: 2.65→2.79 Å / Redundancy: 5.9 % / Rmerge(I) obs: 0.489 / Mean I/σ(I) obs: 3.4 / % possible all: 100 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB entry 1CVU Resolution: 2.65→20 Å / Cor.coef. Fo:Fc: 0.945 / Cor.coef. Fo:Fc free: 0.911 / Occupancy max: 1 / Occupancy min: 0.5 / SU B: 22.664 / SU ML: 0.219 / Cross valid method: THROUGHOUT / ESU R: 1.619 / ESU R Free: 0.312 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: MASK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 38.339 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.348 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.65→20 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.65→2.718 Å / Total num. of bins used: 20
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller


























 PDBj
PDBj








