[English] 日本語
 Yorodumi
Yorodumi- PDB-3e7t: Structure of murine iNOS oxygenase domain with inhibitor AR-C102222 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3e7t | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of murine iNOS oxygenase domain with inhibitor AR-C102222 | |||||||||
 Components Components | Nitric oxide synthase, inducible | |||||||||
 Keywords Keywords | OXIDOREDUCTASE / NITRIC OXIDE SYNTHASE / NOS / HEME / TETRAHYDROBIOPTERIN / OXIDOREDUCTASE Calmodulin-binding / FAD / FMN / Iron / Metal-binding / NADP / Polymorphism / Zinc | |||||||||
| Function / homology |  Function and homology information Function and homology informationNitric oxide stimulates guanylate cyclase / ROS and RNS production in phagocytes / peptidyl-cysteine S-nitrosylation / Peroxisomal protein import / prostaglandin secretion / tetrahydrobiopterin binding / arginine binding / superoxide metabolic process / regulation of cytokine production involved in inflammatory response / Fc-gamma receptor signaling pathway involved in phagocytosis ...Nitric oxide stimulates guanylate cyclase / ROS and RNS production in phagocytes / peptidyl-cysteine S-nitrosylation / Peroxisomal protein import / prostaglandin secretion / tetrahydrobiopterin binding / arginine binding / superoxide metabolic process / regulation of cytokine production involved in inflammatory response / Fc-gamma receptor signaling pathway involved in phagocytosis / cellular response to cytokine stimulus / cortical cytoskeleton / nitric-oxide synthase (NADPH) / nitric-oxide synthase activity / L-arginine catabolic process / nitric oxide biosynthetic process / regulation of insulin secretion / positive regulation of interleukin-8 production / response to bacterium / circadian rhythm / negative regulation of protein catabolic process / positive regulation of interleukin-6 production / cellular response to type II interferon / cellular response to xenobiotic stimulus / peroxisome / FMN binding / NADP binding / flavin adenine dinucleotide binding / regulation of cell population proliferation / cellular response to lipopolysaccharide / response to lipopolysaccharide / response to hypoxia / calmodulin binding / defense response to bacterium / inflammatory response / negative regulation of gene expression / heme binding / perinuclear region of cytoplasm / protein homodimerization activity / metal ion binding / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.6 Å MOLECULAR REPLACEMENT / Resolution: 2.6 Å | |||||||||
 Authors Authors | Garcin, E.D. / Arvai, A.S. / Rosenfeld, R.J. / Kroeger, M.D. / Crane, B.R. / Andersson, G. / Andrews, G. / Hamley, P.J. / Mallinder, P.R. / Nicholls, D.J. ...Garcin, E.D. / Arvai, A.S. / Rosenfeld, R.J. / Kroeger, M.D. / Crane, B.R. / Andersson, G. / Andrews, G. / Hamley, P.J. / Mallinder, P.R. / Nicholls, D.J. / St-Gallay, S.A. / Tinker, A.C. / Gensmantel, N.P. / Mete, A. / Cheshire, D.R. / Connolly, S. / Stuehr, D.J. / Aberg, A. / Wallace, A.V. / Tainer, J.A. / Getzoff, E.D. | |||||||||
 Citation Citation |  Journal: Nat.Chem.Biol. / Year: 2008 Journal: Nat.Chem.Biol. / Year: 2008Title: Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase. Authors: Garcin, E.D. / Arvai, A.S. / Rosenfeld, R.J. / Kroeger, M.D. / Crane, B.R. / Andersson, G. / Andrews, G. / Hamley, P.J. / Mallinder, P.R. / Nicholls, D.J. / St-Gallay, S.A. / Tinker, A.C. / ...Authors: Garcin, E.D. / Arvai, A.S. / Rosenfeld, R.J. / Kroeger, M.D. / Crane, B.R. / Andersson, G. / Andrews, G. / Hamley, P.J. / Mallinder, P.R. / Nicholls, D.J. / St-Gallay, S.A. / Tinker, A.C. / Gensmantel, N.P. / Mete, A. / Cheshire, D.R. / Connolly, S. / Stuehr, D.J. / Aberg, A. / Wallace, A.V. / Tainer, J.A. / Getzoff, E.D. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3e7t.cif.gz 3e7t.cif.gz | 192.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3e7t.ent.gz pdb3e7t.ent.gz | 152.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3e7t.json.gz 3e7t.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/e7/3e7t https://data.pdbj.org/pub/pdb/validation_reports/e7/3e7t ftp://data.pdbj.org/pub/pdb/validation_reports/e7/3e7t ftp://data.pdbj.org/pub/pdb/validation_reports/e7/3e7t | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3e65C  3e67C  3e68C  3e6lC  3e6nC  3e6oC  3e6tC  3e7gC  3e7iC  3e7mC  3e7sC  3eahC  3eaiC  3ebdC  3ebfC  3ej8C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 50120.953 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|
-Non-polymers , 5 types, 331 molecules 

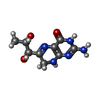






| #2: Chemical | ChemComp-SO4 / | ||||||
|---|---|---|---|---|---|---|---|
| #3: Chemical | | #4: Chemical | #5: Chemical | #6: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.86 Å3/Da / Density % sol: 68.15 % |
|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, hanging drop / pH: 6 Details: LITHIUM SULFATE, MES, BOG, PH 6.0, VAPOR DIFFUSION, HANGING DROP, TEMPERATURE 298K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRL SSRL  / Beamline: BL7-1 / Wavelength: 1.08 / Beamline: BL7-1 / Wavelength: 1.08 |
| Detector | Type: ADSC QUANTUM 4 / Detector: CCD / Date: Feb 16, 1998 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.08 Å / Relative weight: 1 |
| Reflection | Resolution: 2.6→20 Å / Num. obs: 44360 / % possible obs: 96.2 % / Observed criterion σ(I): 0 / Biso Wilson estimate: 42.3 Å2 / Rmerge(I) obs: 0.064 / Rsym value: 0.064 |
| Reflection shell | Resolution: 2.6→2.69 Å / Rmerge(I) obs: 0.407 / Mean I/σ(I) obs: 2.7 / Rsym value: 0.407 / % possible all: 93.6 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 2.6→19.99 Å / Occupancy max: 1 / Occupancy min: 1 / Data cutoff high absF: 415130 / Data cutoff low absF: 0 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: ENGH & HUBER MOLECULAR REPLACEMENT / Resolution: 2.6→19.99 Å / Occupancy max: 1 / Occupancy min: 1 / Data cutoff high absF: 415130 / Data cutoff low absF: 0 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: ENGH & HUBER
| ||||||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 47.03 Å2 / ksol: 0.31 e/Å3 | ||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 144.45 Å2 / Biso mean: 52.166 Å2 / Biso min: 18.53 Å2
| ||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.6→19.99 Å
| ||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.6→2.69 Å / Num. reflection Rwork: 6568 / Total num. of bins used: 6 | ||||||||||||||||||||||||||||
| Xplor file |
|
 Movie
Movie Controller
Controller






















 PDBj
PDBj







