+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: PDB / ID: 3ebf | ||||||
|---|---|---|---|---|---|---|---|
| タイトル | Structure of inhibited murine iNOS oxygenase domain | ||||||
 要素 要素 | Nitric oxide synthase, inducible | ||||||
 キーワード キーワード | OXIDOREDUCTASE / NITRIC OXIDE SYNTHASE / NOS / HEME / TETRAHYDROBIOPTERIN / OXIDOREDUCTASE Calmodulin-binding / FAD / FMN / Iron / Metal-binding / NADP / Polymorphism / Zinc | ||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報Nitric oxide stimulates guanylate cyclase / ROS and RNS production in phagocytes / peptidyl-cysteine S-nitrosylation / Peroxisomal protein import / prostaglandin secretion / tetrahydrobiopterin binding / arginine binding / superoxide metabolic process / regulation of cytokine production involved in inflammatory response / Fc-gamma receptor signaling pathway involved in phagocytosis ...Nitric oxide stimulates guanylate cyclase / ROS and RNS production in phagocytes / peptidyl-cysteine S-nitrosylation / Peroxisomal protein import / prostaglandin secretion / tetrahydrobiopterin binding / arginine binding / superoxide metabolic process / regulation of cytokine production involved in inflammatory response / Fc-gamma receptor signaling pathway involved in phagocytosis / cellular response to cytokine stimulus / cortical cytoskeleton / nitric-oxide synthase (NADPH) / nitric-oxide synthase activity / L-arginine catabolic process / nitric oxide biosynthetic process / regulation of insulin secretion / positive regulation of interleukin-8 production / response to bacterium / circadian rhythm / negative regulation of protein catabolic process / positive regulation of interleukin-6 production / cellular response to type II interferon / cellular response to xenobiotic stimulus / peroxisome / FMN binding / NADP binding / flavin adenine dinucleotide binding / regulation of cell population proliferation / cellular response to lipopolysaccharide / response to lipopolysaccharide / response to hypoxia / calmodulin binding / defense response to bacterium / inflammatory response / negative regulation of gene expression / heme binding / perinuclear region of cytoplasm / protein homodimerization activity / metal ion binding / cytoplasm / cytosol 類似検索 - 分子機能 | ||||||
| 生物種 |  | ||||||
| 手法 |  X線回折 / X線回折 /  シンクロトロン / 解像度: 2.29 Å シンクロトロン / 解像度: 2.29 Å | ||||||
 データ登録者 データ登録者 | Garcin, E.D. / Arvai, A.S. / Rosenfeld, R.J. / Kroeger, M.D. / Crane, B.R. / Andersson, G. / Andrews, G. / Hamley, P.J. / Mallinder, P.R. / Nicholls, D.J. ...Garcin, E.D. / Arvai, A.S. / Rosenfeld, R.J. / Kroeger, M.D. / Crane, B.R. / Andersson, G. / Andrews, G. / Hamley, P.J. / Mallinder, P.R. / Nicholls, D.J. / St-Gallay, S.A. / Tinker, A.C. / Gensmantel, N.P. / Mete, A. / Cheshire, D.R. / Connolly, S. / Stuehr, D.J. / Aberg, A. / Wallace, A.V. / Tainer, J.A. / Getzoff, E.D. | ||||||
 引用 引用 |  ジャーナル: Nat.Chem.Biol. / 年: 2008 ジャーナル: Nat.Chem.Biol. / 年: 2008タイトル: Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase. 著者: Garcin, E.D. / Arvai, A.S. / Rosenfeld, R.J. / Kroeger, M.D. / Crane, B.R. / Andersson, G. / Andrews, G. / Hamley, P.J. / Mallinder, P.R. / Nicholls, D.J. / St-Gallay, S.A. / Tinker, A.C. / ...著者: Garcin, E.D. / Arvai, A.S. / Rosenfeld, R.J. / Kroeger, M.D. / Crane, B.R. / Andersson, G. / Andrews, G. / Hamley, P.J. / Mallinder, P.R. / Nicholls, D.J. / St-Gallay, S.A. / Tinker, A.C. / Gensmantel, N.P. / Mete, A. / Cheshire, D.R. / Connolly, S. / Stuehr, D.J. / Aberg, A. / Wallace, A.V. / Tainer, J.A. / Getzoff, E.D. | ||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 構造ビューア | 分子:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- ダウンロードとリンク
ダウンロードとリンク
- ダウンロード
ダウンロード
| PDBx/mmCIF形式 |  3ebf.cif.gz 3ebf.cif.gz | 208.1 KB | 表示 |  PDBx/mmCIF形式 PDBx/mmCIF形式 |
|---|---|---|---|---|
| PDB形式 |  pdb3ebf.ent.gz pdb3ebf.ent.gz | 163.8 KB | 表示 |  PDB形式 PDB形式 |
| PDBx/mmJSON形式 |  3ebf.json.gz 3ebf.json.gz | ツリー表示 |  PDBx/mmJSON形式 PDBx/mmJSON形式 | |
| その他 |  その他のダウンロード その他のダウンロード |
-検証レポート
| アーカイブディレクトリ |  https://data.pdbj.org/pub/pdb/validation_reports/eb/3ebf https://data.pdbj.org/pub/pdb/validation_reports/eb/3ebf ftp://data.pdbj.org/pub/pdb/validation_reports/eb/3ebf ftp://data.pdbj.org/pub/pdb/validation_reports/eb/3ebf | HTTPS FTP |
|---|
-関連構造データ
| 関連構造データ |  3e65C  3e67C  3e68C  3e6lC  3e6nC  3e6oC  3e6tC  3e7gC  3e7iC  3e7mC  3e7sC  3e7tC  3eahC  3eaiC  3ebdC  3ej8C C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ |
- リンク
リンク
- 集合体
集合体
| 登録構造単位 | 
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||||||||||||||
| 2 | 
| ||||||||||||||||||||||||
| 単位格子 |
| ||||||||||||||||||||||||
| Components on special symmetry positions |
|
- 要素
要素
-タンパク質 , 1種, 2分子 AB
| #1: タンパク質 | 分子量: 50120.953 Da / 分子数: 2 / 由来タイプ: 組換発現 / 由来: (組換発現)   |
|---|
-非ポリマー , 5種, 865分子 

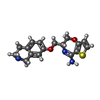






| #2: 化合物 | | #3: 化合物 | #4: 化合物 | #5: 化合物 | ChemComp-SO4 / #6: 水 | ChemComp-HOH / | |
|---|
-実験情報
-実験
| 実験 | 手法:  X線回折 / 使用した結晶の数: 1 X線回折 / 使用した結晶の数: 1 |
|---|
- 試料調製
試料調製
| 結晶 | マシュー密度: 3.81 Å3/Da / 溶媒含有率: 67.68 % |
|---|---|
| 結晶化 | 温度: 298 K / 手法: 蒸気拡散法, ハンギングドロップ法 / pH: 6 詳細: pH 6.0, VAPOR DIFFUSION, HANGING DROP, temperature 298K |
-データ収集
| 放射光源 | 由来:  シンクロトロン / サイト: シンクロトロン / サイト:  SSRL SSRL  / ビームライン: BL9-1 / 波長: 1.08 / ビームライン: BL9-1 / 波長: 1.08 |
|---|---|
| 検出器 | タイプ: ADSC QUANTUM 4 / 検出器: CCD |
| 放射 | プロトコル: SINGLE WAVELENGTH / 単色(M)・ラウエ(L): M / 散乱光タイプ: x-ray |
| 放射波長 | 波長: 1.08 Å / 相対比: 1 |
| 反射 | 解像度: 2.28→31.39 Å / Num. obs: 65348 / Biso Wilson estimate: 29.3 Å2 |
- 解析
解析
| ソフトウェア |
| ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 精密化 | 解像度: 2.29→31.39 Å / Rfactor Rfree error: 0.004 / Occupancy max: 1 / Occupancy min: 0.5 / Data cutoff high absF: 3794811 / Data cutoff low absF: 0 / Isotropic thermal model: RESTRAINED / 交差検証法: THROUGHOUT / σ(F): 0 / 詳細: BULK SOLVENT MODEL USED
| ||||||||||||||||||||||||||||||||||||
| 溶媒の処理 | 溶媒モデル: FLAT MODEL / Bsol: 82.83 Å2 / ksol: 0.4 e/Å3 | ||||||||||||||||||||||||||||||||||||
| 原子変位パラメータ | Biso max: 90 Å2 / Biso mean: 34.718 Å2 / Biso min: 11.69 Å2
| ||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||
| 精密化ステップ | サイクル: LAST / 解像度: 2.29→31.39 Å
| ||||||||||||||||||||||||||||||||||||
| 拘束条件 |
| ||||||||||||||||||||||||||||||||||||
| Xplor file |
|
 ムービー
ムービー コントローラー
コントローラー
















 PDBj
PDBj







