[English] 日本語
 Yorodumi
Yorodumi- PDB-3drc: INVESTIGATION OF THE FUNCTIONAL ROLE OF TRYPTOPHAN-22 IN ESCHERIC... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3drc | ||||||
|---|---|---|---|---|---|---|---|
| Title | INVESTIGATION OF THE FUNCTIONAL ROLE OF TRYPTOPHAN-22 IN ESCHERICHIA COLI DIHYDROFOLATE REDUCTASE BY SITE-DIRECTED MUTAGENESIS | ||||||
 Components Components | DIHYDROFOLATE REDUCTASE | ||||||
 Keywords Keywords | OXIDOREDUCTASE | ||||||
| Function / homology |  Function and homology information Function and homology informationmethotrexate binding / dihydrofolic acid binding / 10-formyltetrahydrofolate biosynthetic process / response to methotrexate / NADP+ binding / folic acid biosynthetic process / folic acid binding / dihydrofolate metabolic process / dihydrofolate reductase / dihydrofolate reductase activity ...methotrexate binding / dihydrofolic acid binding / 10-formyltetrahydrofolate biosynthetic process / response to methotrexate / NADP+ binding / folic acid biosynthetic process / folic acid binding / dihydrofolate metabolic process / dihydrofolate reductase / dihydrofolate reductase activity / folic acid metabolic process / NADPH binding / tetrahydrofolate biosynthetic process / one-carbon metabolic process / NADP binding / response to xenobiotic stimulus / response to antibiotic / cytosol Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / Resolution: 1.9 Å X-RAY DIFFRACTION / Resolution: 1.9 Å | ||||||
 Authors Authors | Oatley, S.J. / Kraut, J. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 1991 Journal: Biochemistry / Year: 1991Title: Investigation of the functional role of tryptophan-22 in Escherichia coli dihydrofolate reductase by site-directed mutagenesis. Authors: Warren, M.S. / Brown, K.A. / Farnum, M.F. / Howell, E.E. / Kraut, J. #1:  Journal: Biochemistry / Year: 1991 Journal: Biochemistry / Year: 1991Title: Crystal Structure of Unliganded Escherichia Coli Dihydrofolate Reductase. Ligand-Induced Conformational Changes and Cooperativity in Binding Authors: Bystroff, C. / Kraut, J. #2:  Journal: Biochemistry / Year: 1990 Journal: Biochemistry / Year: 1990Title: Crystal Structures of Escherichia Coli Dihydrofolate Reductase: The Nadp+ Holoenzyme and the Folate Nadp+ Ternary Complex. Substrate Binding and a Model for the Transition State Authors: Bystroff, C. / Oatley, S.J. / Kraut, J. #3:  Journal: Biochemistry / Year: 1990 Journal: Biochemistry / Year: 1990Title: Crystal Structures of Recombinant Human Dihydrofolate Reductase Complexed with Folate and 5-Deazafolate Authors: Davies II, J.F. / Delcamp, T.J. / Prendergast, N.J. / Ashford, V.A. / Freisheim, J.H. / Kraut, J. #4:  Journal: Science / Year: 1986 Journal: Science / Year: 1986Title: Functional Role of Aspartic Acid-27 in Dihydrofolate Reductase Revealed by Mutagenesis Authors: Howell, E.E. / Villafranca, J.E. / Warren, M.S. / Oatley, S.J. / Kraut, J. #5:  Journal: J.Biol.Chem. / Year: 1982 Journal: J.Biol.Chem. / Year: 1982Title: Crystal Structures of Escherichia Coli and Lactobacillus Casei Dihydrofolate Reductase Refined at 1.7 Angstroms Resolution. I. General Features and Binding of Methotrexate Authors: Bolin, J.T. / Filman, D.J. / Matthews, D.A. / Hamlin, R.C. / Kraut, J. #6:  Journal: J.Biol.Chem. / Year: 1982 Journal: J.Biol.Chem. / Year: 1982Title: Crystal Structures of Escherichia Coli and Lactobacillus Casei Dihydrofolate Reductase Refined at 1.7 Angstroms Resolution. II. Environment of Bound Nadph and Implications for Catalysis Authors: Filman, D.J. / Bolin, J.T. / Matthews, D.A. / Kraut, J. | ||||||
| History |
|
- Structure visualization
Structure visualization
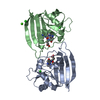
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3drc.cif.gz 3drc.cif.gz | 83.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3drc.ent.gz pdb3drc.ent.gz | 61.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3drc.json.gz 3drc.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  3drc_validation.pdf.gz 3drc_validation.pdf.gz | 502.9 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  3drc_full_validation.pdf.gz 3drc_full_validation.pdf.gz | 522.2 KB | Display | |
| Data in XML |  3drc_validation.xml.gz 3drc_validation.xml.gz | 11.5 KB | Display | |
| Data in CIF |  3drc_validation.cif.gz 3drc_validation.cif.gz | 17.8 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/dr/3drc https://data.pdbj.org/pub/pdb/validation_reports/dr/3drc ftp://data.pdbj.org/pub/pdb/validation_reports/dr/3drc ftp://data.pdbj.org/pub/pdb/validation_reports/dr/3drc | HTTPS FTP |
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Atom site foot note | 1: GLY A 95 - GLY A 96 OMEGA ANGLE = 3.516 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION 2: GLY B 95 - GLY B 96 OMEGA ANGLE = 4.003 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION | ||||||||
| Noncrystallographic symmetry (NCS) | NCS oper: (Code: given Matrix: (-0.92389, -0.37481, 0.07712), Vector: Details | THE TRANSFORMATION PRESENTED ON *MTRIX* RECORDS BELOW WILL YIELD APPROXIMATE COORDINATES FOR CHAIN *A* WHEN APPLIED TO CHAIN *B*. | |
- Components
Components
| #1: Protein | Mass: 18020.326 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  #2: Chemical | #3: Chemical | #4: Chemical | ChemComp-CA / | #5: Water | ChemComp-HOH / | Sequence details | RESIDUE 154 IS GLU IN THE RT500 STRAIN. IN THE MB1428 STRAIN (ENTRY 4DFR), THIS RESIDUE IS A LYS. | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.56 Å3/Da / Density % sol: 52.03 % | |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | *PLUS Temperature: 4 ℃ / Method: vapor diffusion | |||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Reflection | *PLUS Highest resolution: 1.8 Å / % possible obs: 96 % / Num. measured all: 30609 / Rmerge(I) obs: 0.049 |
|---|
- Processing
Processing
| Software | Name: TNT / Classification: refinement | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Rfactor obs: 0.156 / Highest resolution: 1.9 Å Details: MOLECULE DESIGNATED AS CHAIN B BELOW IS PREFERRED FOR STRUCTURAL COMPARISONS BECAUSE IT IS MORE COMPLETE AND LESS PERTURBED BY INTERMOLECULAR CONTACTS. | ||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Highest resolution: 1.9 Å
| ||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||
| Software | *PLUS Name: TNT / Classification: refinement | ||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS Type: t_angle_d / Dev ideal: 2.99 |
 Movie
Movie Controller
Controller

















 PDBj
PDBj