[English] 日本語
 Yorodumi
Yorodumi- PDB-3bdx: Crystal structure of the unstable and highly fibrillogenic Pro7Se... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3bdx | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of the unstable and highly fibrillogenic Pro7Ser mutant of the Recombinant variable domain 6AJL2 | ||||||
 Components Components | Amyloid lambda 6 light chain variable region PIP (fragment) | ||||||
 Keywords Keywords | IMMUNE SYSTEM / Lambda VI subgroup / light chain variable domain / beta-sandwich / immunoglobulin / AL amyloidosis | ||||||
| Function / homology |  Function and homology information Function and homology information | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.3 Å MOLECULAR REPLACEMENT / Resolution: 2.3 Å | ||||||
 Authors Authors | Hernandez-Santoyo, A. / Fuentes-Silva, D. / Del Pozo Yauner, L. / Becerril, B. / Rodriguez-Romero, A. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2010 Journal: J.Mol.Biol. / Year: 2010Title: A single mutation at the sheet switch region results in conformational changes favoring lambda6 light-chain fibrillogenesis. Authors: Hernandez-Santoyo, A. / del Pozo Yauner, L. / Fuentes-Silva, D. / Ortiz, E. / Rudino-Pinera, E. / Sanchez-Lopez, R. / Horjales, E. / Becerril, B. / Rodriguez-Romero, A. | ||||||
| History |
| ||||||
| Remark 999 | SEQUENCE THE RESIDUES ARE NUMBERED CONSECUTIVELY IN THE POLYPEPTIDE CHAIN AND DO NOT FOLLOW THE ... SEQUENCE THE RESIDUES ARE NUMBERED CONSECUTIVELY IN THE POLYPEPTIDE CHAIN AND DO NOT FOLLOW THE KABAT NUMBERING SCHEME. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3bdx.cif.gz 3bdx.cif.gz | 77.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3bdx.ent.gz pdb3bdx.ent.gz | 59 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3bdx.json.gz 3bdx.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  3bdx_validation.pdf.gz 3bdx_validation.pdf.gz | 482.9 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  3bdx_full_validation.pdf.gz 3bdx_full_validation.pdf.gz | 488.4 KB | Display | |
| Data in XML |  3bdx_validation.xml.gz 3bdx_validation.xml.gz | 16 KB | Display | |
| Data in CIF |  3bdx_validation.cif.gz 3bdx_validation.cif.gz | 21.3 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/bd/3bdx https://data.pdbj.org/pub/pdb/validation_reports/bd/3bdx ftp://data.pdbj.org/pub/pdb/validation_reports/bd/3bdx ftp://data.pdbj.org/pub/pdb/validation_reports/bd/3bdx | HTTPS FTP |
-Related structure data
| Related structure data |  2w0kC  3b5gSC S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| 2 | 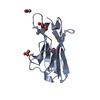
| |||||||||
| 3 | 
| |||||||||
| 4 | 
| |||||||||
| Unit cell |
| |||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Antibody | Mass: 11951.873 Da / Num. of mol.: 3 / Fragment: Residues 1-111 / Mutation: P7S Source method: isolated from a genetically manipulated source Details: Promotor: lac Resistence ampicillin / Source: (gene. exp.)  Homo sapiens (human) / Strain: Lambda VI light chain subgroup / Gene: VL gene segment 6a and JL2/3 gene segment / Plasmid: pSyn1 / Production host: Homo sapiens (human) / Strain: Lambda VI light chain subgroup / Gene: VL gene segment 6a and JL2/3 gene segment / Plasmid: pSyn1 / Production host:  #2: Chemical | ChemComp-ACT / #3: Chemical | ChemComp-GOL / #4: Chemical | ChemComp-MES / | #5: Water | ChemComp-HOH / | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.77 Å3/Da / Density % sol: 55.57 % |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, hanging drop / pH: 6.5 Details: Drops consisted of 5 microliters of protein solution (7 mg/mL) plus 5 microliters of 0.1 M MES pH 6.5, 2.0 M Sodium acetate trihydrate, VAPOR DIFFUSION, HANGING DROP, temperature 291K |
-Data collection
| Diffraction | Mean temperature: 103 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 Å ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 Å |
| Detector | Type: RIGAKU RAXIS IIC / Detector: IMAGE PLATE / Date: Sep 7, 2005 / Details: Mirrors |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2.3→29.92 Å / Num. obs: 16902 / % possible obs: 93.9 % / Redundancy: 2.3 % / Biso Wilson estimate: 14.9 Å2 / Rmerge(I) obs: 0.084 / Net I/σ(I): 8.7 |
| Reflection shell | Resolution: 2.3→2.44 Å / Redundancy: 2.2 % / Rmerge(I) obs: 0.227 / Mean I/σ(I) obs: 3.9 / Num. unique all: 2544 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB entry 3B5G Resolution: 2.3→29.92 Å / Rfactor Rfree error: 0.006 / Data cutoff high absF: 2014859.88 / Data cutoff low absF: 0 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: Engh & Huber
| ||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 30.4523 Å2 / ksol: 0.39712 e/Å3 | ||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 22.3 Å2
| ||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.3→29.92 Å
| ||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.3→2.44 Å / Rfactor Rfree error: 0.018 / Total num. of bins used: 6
| ||||||||||||||||||||||||||||||||||||
| Xplor file |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj








