[English] 日本語
 Yorodumi
Yorodumi- PDB-329d: EFFECT OF CYTOSINE METHYLATION ON DNA-DNA RECOGNITION AT CPG STEPS -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 329d | ||||||
|---|---|---|---|---|---|---|---|
| Title | EFFECT OF CYTOSINE METHYLATION ON DNA-DNA RECOGNITION AT CPG STEPS | ||||||
 Components Components |
| ||||||
 Keywords Keywords | DNA / B-DNA / DOUBLE HELIX | ||||||
| Function / homology | DNA / DNA (> 10) Function and homology information Function and homology information | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.7 Å MOLECULAR REPLACEMENT / Resolution: 2.7 Å | ||||||
 Authors Authors | Mayer-Jung, C. / Moras, D. / Timsit, Y. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1997 Journal: J.Mol.Biol. / Year: 1997Title: Effect of cytosine methylation on DNA-DNA recognition at CpG steps. Authors: Mayer-Jung, C. / Moras, D. / Timsit, Y. #1:  Journal: J.Mol.Biol. / Year: 1995 Journal: J.Mol.Biol. / Year: 1995Title: Self-Fitting and Self-Modifying Properties of the B-DNA Molecule Authors: Timsit, Y. / Moras, D. #2:  Journal: Nature / Year: 1991 Journal: Nature / Year: 1991Title: Base-Pairing Shift in the Major Groove of (CA)n Tracts by B-DNA Crystal Structures Authors: Timsit, Y. / Vilbois, E. / Moras, D. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  329d.cif.gz 329d.cif.gz | 23 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb329d.ent.gz pdb329d.ent.gz | 15.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  329d.json.gz 329d.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/29/329d https://data.pdbj.org/pub/pdb/validation_reports/29/329d ftp://data.pdbj.org/pub/pdb/validation_reports/29/329d ftp://data.pdbj.org/pub/pdb/validation_reports/29/329d | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 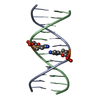
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: DNA chain | Mass: 3623.372 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source |
|---|---|
| #2: DNA chain | Mass: 3734.430 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source |
| #3: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.6 Å3/Da / Density % sol: 52.75 % | ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Method: vapor diffusion / pH: 6 / Details: pH 6.00, VAPOR DIFFUSION | ||||||||||||||||||||||||||||||||||||||||
| Components of the solutions |
| ||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 15 ℃ / pH: 6 / Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 273 K |
|---|---|
| Detector | Type: SIEMENS / Detector: AREA DETECTOR / Date: Mar 1, 1993 |
| Radiation | Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Relative weight: 1 |
| Reflection | Resolution: 2.7→19.5 Å / Num. obs: 1789 / % possible obs: 95 % / Observed criterion σ(I): 1 / Redundancy: 8.2 % / Rmerge(I) obs: 0.054 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: BDL035 Resolution: 2.7→8 Å / Data cutoff high absF: 1000 / Data cutoff low absF: 0.001 / σ(F): 2 /
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 36 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine Biso |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.7→8 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Classification: refinement X-PLOR / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 2.7 Å / Lowest resolution: 8 Å / σ(F): 2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS Biso mean: 36 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller
















 PDBj
PDBj







































