Entry Database : PDB / ID : 2zeqTitle Crystal structure of ubiquitin-like domain of murine Parkin E3 ubiquitin-protein ligase parkin Keywords / / / / / / / / / / / / / / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Mus musculus (house mouse)Method / / Resolution : 1.65 Å Authors Tomoo, K. Journal : Biochim.Biophys.Acta / Year : 2008Title : Crystal structure and molecular dynamics simulation of ubiquitin-like domain of murine parkinAuthors : Tomoo, K. / Mukai, Y. / In, Y. / Miyagawa, H. / Kitamura, K. / Yamano, A. / Shindo, H. / Ishida, T. History Deposition Dec 14, 2007 Deposition site / Processing site Revision 1.0 Aug 19, 2008 Provider / Type Revision 1.1 Jul 13, 2011 Group Revision 1.2 Mar 13, 2024 Group / Database referencesCategory chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / struct_ref_seq_dif Item / _database_2.pdbx_database_accession / _struct_ref_seq_dif.details
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SAD / Resolution: 1.65 Å
SAD / Resolution: 1.65 Å  Authors
Authors Citation
Citation Journal: Biochim.Biophys.Acta / Year: 2008
Journal: Biochim.Biophys.Acta / Year: 2008 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 2zeq.cif.gz
2zeq.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb2zeq.ent.gz
pdb2zeq.ent.gz PDB format
PDB format 2zeq.json.gz
2zeq.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads 2zeq_validation.pdf.gz
2zeq_validation.pdf.gz wwPDB validaton report
wwPDB validaton report 2zeq_full_validation.pdf.gz
2zeq_full_validation.pdf.gz 2zeq_validation.xml.gz
2zeq_validation.xml.gz 2zeq_validation.cif.gz
2zeq_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/ze/2zeq
https://data.pdbj.org/pub/pdb/validation_reports/ze/2zeq ftp://data.pdbj.org/pub/pdb/validation_reports/ze/2zeq
ftp://data.pdbj.org/pub/pdb/validation_reports/ze/2zeq Links
Links Assembly
Assembly
 Components
Components

 X-RAY DIFFRACTION / Number of used crystals: 2
X-RAY DIFFRACTION / Number of used crystals: 2  Sample preparation
Sample preparation ROTATING ANODE / Type: RIGAKU FR-E+ DW / Wavelength: 1.5418 Å
ROTATING ANODE / Type: RIGAKU FR-E+ DW / Wavelength: 1.5418 Å Processing
Processing SAD / Resolution: 1.65→39.89 Å / Cross valid method: THROUGHOUT
SAD / Resolution: 1.65→39.89 Å / Cross valid method: THROUGHOUT Movie
Movie Controller
Controller



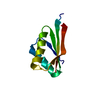









 PDBj
PDBj



