[English] 日本語
 Yorodumi
Yorodumi- PDB-2vnt: Urokinase-Type Plasminogen Activator Inhibitor Complex with a 1-(... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2vnt | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Urokinase-Type Plasminogen Activator Inhibitor Complex with a 1-(7- SULPHOAMIDOISOQUINOLINYL)GUANIDINE | |||||||||
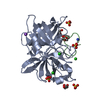
 Components Components | UROKINASE-TYPE PLASMINOGEN ACTIVATOR | |||||||||
 Keywords Keywords | HYDROLASE / UPA / INHIBITOR COMPLEX | |||||||||
| Function / homology |  Function and homology information Function and homology informationu-plasminogen activator / regulation of smooth muscle cell-matrix adhesion / regulation of integrin-mediated signaling pathway / urokinase plasminogen activator signaling pathway / regulation of plasminogen activation / regulation of fibrinolysis / protein complex involved in cell-matrix adhesion / regulation of wound healing / negative regulation of plasminogen activation / serine-type endopeptidase complex ...u-plasminogen activator / regulation of smooth muscle cell-matrix adhesion / regulation of integrin-mediated signaling pathway / urokinase plasminogen activator signaling pathway / regulation of plasminogen activation / regulation of fibrinolysis / protein complex involved in cell-matrix adhesion / regulation of wound healing / negative regulation of plasminogen activation / serine-type endopeptidase complex / regulation of smooth muscle cell migration / Dissolution of Fibrin Clot / smooth muscle cell migration / plasminogen activation / regulation of cell adhesion mediated by integrin / tertiary granule membrane / negative regulation of fibrinolysis / regulation of cell adhesion / positive regulation of epidermal growth factor receptor signaling pathway / specific granule membrane / serine protease inhibitor complex / fibrinolysis / chemotaxis / blood coagulation / regulation of cell population proliferation / response to hypoxia / positive regulation of cell migration / receptor ligand activity / external side of plasma membrane / serine-type endopeptidase activity / focal adhesion / Neutrophil degranulation / cell surface / signal transduction / proteolysis / extracellular space / extracellular exosome / extracellular region / plasma membrane Similarity search - Function | |||||||||
| Biological species |  HOMO SAPIENS (human) HOMO SAPIENS (human) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / OTHER / Resolution: 2.2 Å SYNCHROTRON / OTHER / Resolution: 2.2 Å | |||||||||
 Authors Authors | Fish, P.V. / Barber, C.G. / Brown, D.G. / Butt, R. / Henry, B.T. / Horne, V. / Huggins, J.P. / McCleverty, D. / Phillips, C. / Webster, R. ...Fish, P.V. / Barber, C.G. / Brown, D.G. / Butt, R. / Henry, B.T. / Horne, V. / Huggins, J.P. / McCleverty, D. / Phillips, C. / Webster, R. / Dickinson, R.P. / Collis, M.G. / King, E. / O'Gara, M. / McIntosh, F. | |||||||||
 Citation Citation |  Journal: J.Med.Chem. / Year: 2007 Journal: J.Med.Chem. / Year: 2007Title: Selective Urokinase-Type Plasminogen Activator (Upa) Inhibitors 4. 1-(7-Sulphonamidoisoquinolinyl) Guanidines Authors: Fish, P.V. / Barber, C.G. / Brown, D.G. / Butt, R. / Henry, B.T. / Horne, V. / Huggins, J.P. / McCleverty, D. / Phillips, C. / Webster, R. / Dickinson, R.P. / Collis, M.G. / King, E. / O'Gara, M. / McIntosh, F. | |||||||||
| History |
| |||||||||
| Remark 700 | SHEET DETERMINATION METHOD: DSSP THE SHEETS PRESENTED AS "AA" IN EACH CHAIN ON SHEET RECORDS BELOW ... SHEET DETERMINATION METHOD: DSSP THE SHEETS PRESENTED AS "AA" IN EACH CHAIN ON SHEET RECORDS BELOW IS ACTUALLY AN 8-STRANDED BARREL THIS IS REPRESENTED BY A 9-STRANDED SHEET IN WHICH THE FIRST AND LAST STRANDS ARE IDENTICAL. THE SHEETS PRESENTED AS "AB" IN EACH CHAIN ON SHEET RECORDS BELOW IS ACTUALLY AN 7-STRANDED BARREL THIS IS REPRESENTED BY A 8-STRANDED SHEET IN WHICH THE FIRST AND LAST STRANDS ARE IDENTICAL. THE SHEETS PRESENTED AS "BA" IN EACH CHAIN ON SHEET RECORDS BELOW IS ACTUALLY AN 7-STRANDED BARREL THIS IS REPRESENTED BY A 8-STRANDED SHEET IN WHICH THE FIRST AND LAST STRANDS ARE IDENTICAL. THE SHEETS PRESENTED AS "BB" IN EACH CHAIN ON SHEET RECORDS BELOW IS ACTUALLY AN 5-STRANDED BARREL THIS IS REPRESENTED BY A 6-STRANDED SHEET IN WHICH THE FIRST AND LAST STRANDS ARE IDENTICAL. THE SHEETS PRESENTED AS "CA" IN EACH CHAIN ON SHEET RECORDS BELOW IS ACTUALLY AN 6-STRANDED BARREL THIS IS REPRESENTED BY A 7-STRANDED SHEET IN WHICH THE FIRST AND LAST STRANDS ARE IDENTICAL. THE SHEETS PRESENTED AS "CB" IN EACH CHAIN ON SHEET RECORDS BELOW IS ACTUALLY AN 5-STRANDED BARREL THIS IS REPRESENTED BY A 6-STRANDED SHEET IN WHICH THE FIRST AND LAST STRANDS ARE IDENTICAL. THE SHEETS PRESENTED AS "DA" IN EACH CHAIN ON SHEET RECORDS BELOW IS ACTUALLY AN 6-STRANDED BARREL THIS IS REPRESENTED BY A 7-STRANDED SHEET IN WHICH THE FIRST AND LAST STRANDS ARE IDENTICAL. THE SHEETS PRESENTED AS "DB" IN EACH CHAIN ON SHEET RECORDS BELOW IS ACTUALLY AN 5-STRANDED BARREL THIS IS REPRESENTED BY A 6-STRANDED SHEET IN WHICH THE FIRST AND LAST STRANDS ARE IDENTICAL. THE SHEETS PRESENTED AS "EA" IN EACH CHAIN ON SHEET RECORDS BELOW IS ACTUALLY AN 6-STRANDED BARREL THIS IS REPRESENTED BY A 7-STRANDED SHEET IN WHICH THE FIRST AND LAST STRANDS ARE IDENTICAL. THE SHEETS PRESENTED AS "EB" IN EACH CHAIN ON SHEET RECORDS BELOW IS ACTUALLY AN 5-STRANDED BARREL THIS IS REPRESENTED BY A 6-STRANDED SHEET IN WHICH THE FIRST AND LAST STRANDS ARE IDENTICAL. THE SHEETS PRESENTED AS "FA" IN EACH CHAIN ON SHEET RECORDS BELOW IS ACTUALLY AN 6-STRANDED BARREL THIS IS REPRESENTED BY A 7-STRANDED SHEET IN WHICH THE FIRST AND LAST STRANDS ARE IDENTICAL. THE SHEETS PRESENTED AS "FB" IN EACH CHAIN ON SHEET RECORDS BELOW IS ACTUALLY AN 5-STRANDED BARREL THIS IS REPRESENTED BY A 6-STRANDED SHEET IN WHICH THE FIRST AND LAST STRANDS ARE IDENTICAL. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2vnt.cif.gz 2vnt.cif.gz | 329.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2vnt.ent.gz pdb2vnt.ent.gz | 269.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2vnt.json.gz 2vnt.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/vn/2vnt https://data.pdbj.org/pub/pdb/validation_reports/vn/2vnt ftp://data.pdbj.org/pub/pdb/validation_reports/vn/2vnt ftp://data.pdbj.org/pub/pdb/validation_reports/vn/2vnt | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||||||||||||||||||||||||||||||||
| 2 | 
| ||||||||||||||||||||||||||||||||||||||||||
| 3 | 
| ||||||||||||||||||||||||||||||||||||||||||
| 4 | 
| ||||||||||||||||||||||||||||||||||||||||||
| 5 | 
| ||||||||||||||||||||||||||||||||||||||||||
| 6 | 
| ||||||||||||||||||||||||||||||||||||||||||
| Unit cell |
| ||||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments: Component-ID: 1 / Ens-ID: 1 / Beg auth comp-ID: CYS / Beg label comp-ID: CYS / End auth comp-ID: GLU / End label comp-ID: GLU / Refine code: 4 / Auth seq-ID: 1 - 257 / Label seq-ID: 13 - 271
NCS oper:
|
- Components
Components
| #1: Protein | Mass: 31168.613 Da / Num. of mol.: 6 / Fragment: CATALYTIC DOMAIN, RESIDUES 156-431 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  HOMO SAPIENS (human) / Production host: HOMO SAPIENS (human) / Production host:  #2: Chemical | ChemComp-QGG / #3: Chemical | ChemComp-SO4 / #4: Water | ChemComp-HOH / | Has protein modification | Y | Nonpolymer details | UNKNOWN (QGG): 1- 7-SULPHOAMID | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.63 Å3/Da / Density % sol: 53.28 % / Description: NONE |
|---|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SRS SRS  / Type: / Type:  SRS SRS  / Wavelength: 1 / Wavelength: 1 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 2.2→30 Å / Num. obs: 97771 / % possible obs: 99.5 % / Observed criterion σ(I): 0 / Redundancy: 3.3 % / Rmerge(I) obs: 0.08 |
- Processing
Processing
| Software | Name: REFMAC / Version: 5.2.0019 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure: OTHER Starting model: NONE Resolution: 2.2→35 Å / Cor.coef. Fo:Fc: 0.914 / Cor.coef. Fo:Fc free: 0.859 / Cross valid method: THROUGHOUT / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 35.55 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.2→35 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller




























































 PDBj
PDBj