[English] 日本語
 Yorodumi
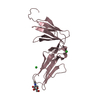
Yorodumi- PDB-2lvp: gp78CUE domain bound to the distal ubiquitin of K48-linked diubiquitin -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2lvp | ||||||
|---|---|---|---|---|---|---|---|
| Title | gp78CUE domain bound to the distal ubiquitin of K48-linked diubiquitin | ||||||
 Components Components |
| ||||||
 Keywords Keywords | SIGNALING PROTEIN/LIGASE / CUE domain / SIGNALING PROTEIN-LIGASE complex | ||||||
| Function / homology |  Function and homology information Function and homology informationregulation of SREBP signaling pathway / RING-type E3 ubiquitin transferase (cysteine targeting) / endoplasmic reticulum mannose trimming / protein K27-linked ubiquitination / endoplasmic reticulum quality control compartment / BAT3 complex binding / Derlin-1 retrotranslocation complex / non-canonical NF-kappaB signal transduction / ubiquitin-ubiquitin ligase activity / ubiquitin-specific protease binding ...regulation of SREBP signaling pathway / RING-type E3 ubiquitin transferase (cysteine targeting) / endoplasmic reticulum mannose trimming / protein K27-linked ubiquitination / endoplasmic reticulum quality control compartment / BAT3 complex binding / Derlin-1 retrotranslocation complex / non-canonical NF-kappaB signal transduction / ubiquitin-ubiquitin ligase activity / ubiquitin-specific protease binding / ubiquitin ligase complex / protein K48-linked ubiquitination / endoplasmic reticulum unfolded protein response / protein autoubiquitination / ERAD pathway / Maturation of protein E / Maturation of protein E / ER Quality Control Compartment (ERQC) / Myoclonic epilepsy of Lafora / FLT3 signaling by CBL mutants / Constitutive Signaling by NOTCH1 HD Domain Mutants / IRAK2 mediated activation of TAK1 complex / Prevention of phagosomal-lysosomal fusion / Alpha-protein kinase 1 signaling pathway / Glycogen synthesis / IRAK1 recruits IKK complex / IRAK1 recruits IKK complex upon TLR7/8 or 9 stimulation / Endosomal Sorting Complex Required For Transport (ESCRT) / Membrane binding and targetting of GAG proteins / Negative regulation of FLT3 / Regulation of TBK1, IKKε (IKBKE)-mediated activation of IRF3, IRF7 / PTK6 Regulates RTKs and Their Effectors AKT1 and DOK1 / Regulation of TBK1, IKKε-mediated activation of IRF3, IRF7 upon TLR3 ligation / IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation / NOTCH2 Activation and Transmission of Signal to the Nucleus / TICAM1,TRAF6-dependent induction of TAK1 complex / TICAM1-dependent activation of IRF3/IRF7 / APC/C:Cdc20 mediated degradation of Cyclin B / Regulation of FZD by ubiquitination / Downregulation of ERBB4 signaling / APC-Cdc20 mediated degradation of Nek2A / p75NTR recruits signalling complexes / InlA-mediated entry of Listeria monocytogenes into host cells / TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling / TRAF6-mediated induction of TAK1 complex within TLR4 complex / Regulation of pyruvate metabolism / NF-kB is activated and signals survival / Regulation of innate immune responses to cytosolic DNA / Pexophagy / Downregulation of ERBB2:ERBB3 signaling / NRIF signals cell death from the nucleus / Activated NOTCH1 Transmits Signal to the Nucleus / Regulation of PTEN localization / VLDLR internalisation and degradation / Regulation of BACH1 activity / Synthesis of active ubiquitin: roles of E1 and E2 enzymes / MAP3K8 (TPL2)-dependent MAPK1/3 activation / ubiquitin binding / Translesion synthesis by REV1 / TICAM1, RIP1-mediated IKK complex recruitment / Translesion synthesis by POLK / InlB-mediated entry of Listeria monocytogenes into host cell / Activation of IRF3, IRF7 mediated by TBK1, IKKε (IKBKE) / JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 / Josephin domain DUBs / Downregulation of TGF-beta receptor signaling / Translesion synthesis by POLI / Gap-filling DNA repair synthesis and ligation in GG-NER / IKK complex recruitment mediated by RIP1 / Regulation of activated PAK-2p34 by proteasome mediated degradation / PINK1-PRKN Mediated Mitophagy / TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) / TNFR1-induced NF-kappa-B signaling pathway / Autodegradation of Cdh1 by Cdh1:APC/C / TCF dependent signaling in response to WNT / APC/C:Cdc20 mediated degradation of Securin / Regulation of NF-kappa B signaling / N-glycan trimming in the ER and Calnexin/Calreticulin cycle / activated TAK1 mediates p38 MAPK activation / Asymmetric localization of PCP proteins / SCF-beta-TrCP mediated degradation of Emi1 / NIK-->noncanonical NF-kB signaling / Ubiquitin-dependent degradation of Cyclin D / TNFR2 non-canonical NF-kB pathway / Regulation of signaling by CBL / AUF1 (hnRNP D0) binds and destabilizes mRNA / NOTCH3 Activation and Transmission of Signal to the Nucleus / Negative regulators of DDX58/IFIH1 signaling / Vpu mediated degradation of CD4 / Assembly of the pre-replicative complex / Ubiquitin-Mediated Degradation of Phosphorylated Cdc25A / Deactivation of the beta-catenin transactivating complex / Negative regulation of FGFR3 signaling / Peroxisomal protein import / Fanconi Anemia Pathway / Degradation of DVL / Dectin-1 mediated noncanonical NF-kB signaling / Cdc20:Phospho-APC/C mediated degradation of Cyclin A / Stabilization of p53 / Negative regulation of FGFR2 signaling Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method | SOLUTION NMR / simulated annealing | ||||||
| Model details | lowest energy, model 1 | ||||||
 Authors Authors | Liu, S. / Chen, Y. / Huang, T. / Tarasov, S.G. / King, A. / Li, J. / Weissman, A.M. / Byrd, R.A. / Das, R. | ||||||
 Citation Citation |  Journal: Structure / Year: 2012 Journal: Structure / Year: 2012Title: Promiscuous Interactions of gp78 E3 Ligase CUE Domain with Polyubiquitin Chains. Authors: Liu, S. / Chen, Y. / Li, J. / Huang, T. / Tarasov, S. / King, A. / Weissman, A.M. / Byrd, R.A. / Das, R. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2lvp.cif.gz 2lvp.cif.gz | 1.2 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2lvp.ent.gz pdb2lvp.ent.gz | 1.1 MB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2lvp.json.gz 2lvp.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  2lvp_validation.pdf.gz 2lvp_validation.pdf.gz | 1.8 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  2lvp_full_validation.pdf.gz 2lvp_full_validation.pdf.gz | 34.4 MB | Display | |
| Data in XML |  2lvp_validation.xml.gz 2lvp_validation.xml.gz | 2.2 MB | Display | |
| Data in CIF |  2lvp_validation.cif.gz 2lvp_validation.cif.gz | 2.5 MB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/lv/2lvp https://data.pdbj.org/pub/pdb/validation_reports/lv/2lvp ftp://data.pdbj.org/pub/pdb/validation_reports/lv/2lvp ftp://data.pdbj.org/pub/pdb/validation_reports/lv/2lvp | HTTPS FTP |
-Related structure data
| Related structure data |  2lvnC  2lvoC  2lvqC C: citing same article ( |
|---|---|
| Similar structure data | |
| Other databases |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 8576.831 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: UBC / Production host: Homo sapiens (human) / Gene: UBC / Production host:  #2: Protein | | Mass: 5966.752 Da / Num. of mol.: 1 / Fragment: CUE domain residues 453-504 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: AMFR, RNF45 / Production host: Homo sapiens (human) / Gene: AMFR, RNF45 / Production host:  References: UniProt: Q9UKV5, Ligases; Forming carbon-nitrogen bonds; Acid-amino-acid ligases (peptide synthases) |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR Details: Amide shifts of gp78CUE and distal Ubiquitin in the gp78CUE/K48-Ub2 complex | ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Contents: 50 mM TRIS, 50 mM sodium chloride, 90% H2O/10% D2O Solvent system: 90% H2O/10% D2O | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| |||||||||
| Sample conditions | pH: 7.2 / Pressure: ambient / Temperature: 298 K |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing / Software ordinal: 1 / Details: FINAL STEP IN WATER | ||||||||||||||||||
| NMR constraints | NOE constraints total: 43 / NOE long range total count: 43 | ||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 200 / Conformers submitted total number: 20 / Maximum lower distance constraint violation: 1.8 Å / Maximum upper distance constraint violation: 8 Å | ||||||||||||||||||
| NMR ensemble rms | Distance rms dev: 0 Å |
 Movie
Movie Controller
Controller











 PDBj
PDBj
















 HSQC
HSQC