[English] 日本語
 Yorodumi
Yorodumi- PDB-2hz8: QM/MM structure refined from NMR-structure of a single chain diir... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2hz8 | ||||||
|---|---|---|---|---|---|---|---|
| Title | QM/MM structure refined from NMR-structure of a single chain diiron protein | ||||||
 Components Components | De novo designed diiron protein | ||||||
 Keywords Keywords | DE NOVO PROTEIN / four-helix bundle | ||||||
| Function / homology | IL-4 antagonist (De novo design) like domain / Four Helix Bundle (Hemerythrin (Met), subunit A) / Up-down Bundle / Mainly Alpha Function and homology information Function and homology information | ||||||
| Biological species |  | ||||||
| Method | SOLUTION NMR / 1. Step: Classical Molecular dynamics simulation starting from NMR-structure, using non-bonded model for metal-site. 2. step: QM, MM MD for relaxation of local frustrations at the metal site. | ||||||
 Authors Authors | Calhoun, J.R. / Liu, W. / Spiegel, K. / Dal Peraro, M. / Klein, M.L. / Wand, A.J. / DeGrado, W.F. | ||||||
 Citation Citation |  Journal: Structure / Year: 2008 Journal: Structure / Year: 2008Title: Solution NMR structure of a designed metalloprotein and complementary molecular dynamics refinement. Authors: Calhoun, J.R. / Liu, W. / Spiegel, K. / Dal Peraro, M. / Klein, M.L. / Valentine, K.G. / Wand, A.J. / Degrado, W.F. #1: Journal: J.Mol.Biol. / Year: 2003 Title: Computational design and characterization of a monomeric helical dinuclear metalloprotein. Authors: Calhoun, J.R. / Kono, H. / Lahr, S. / Wang, W. / DeGrado, W.F. / Saven, J.G. #2: Journal: Biopolymers / Year: 2005 Title: Artificial diiron proteins: from structure to function. Authors: Calhoun, J.R. / Nastri, F. / Magloi, O. / Pavone, V. / Lombardi, A. / DeGrado, W.F. | ||||||
| History |
| ||||||
| Remark 999 | Sequence No suitable database references were found at time of processing |
- Structure visualization
Structure visualization
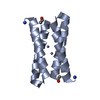
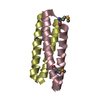
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2hz8.cif.gz 2hz8.cif.gz | 50.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2hz8.ent.gz pdb2hz8.ent.gz | 36.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2hz8.json.gz 2hz8.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/hz/2hz8 https://data.pdbj.org/pub/pdb/validation_reports/hz/2hz8 ftp://data.pdbj.org/pub/pdb/validation_reports/hz/2hz8 ftp://data.pdbj.org/pub/pdb/validation_reports/hz/2hz8 | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 13516.417 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   | ||
|---|---|---|---|
| #2: Chemical | | #3: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||||||||||
| NMR details | Text: The structure was first determined using triple-resonance spectrscopy. The structure was then further refined using classical MD followed by 5 ps of Car Parrinello hybrid QM/MM dynamics. |
- Sample preparation
Sample preparation
| Details |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions | Ionic strength: 10 mM NaCl / pH: 6 / Pressure: ambient / Temperature: 308 K |
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Radiation wavelength | Relative weight: 1 | |||||||||||||||
| NMR spectrometer |
|
- Processing
Processing
| NMR software |
|
|---|
 Movie
Movie Controller
Controller
















 PDBj
PDBj



 HSQC
HSQC