[English] 日本語
 Yorodumi
Yorodumi- PDB-2cnj: NMR studies on the interaction of Insulin-Growth Factor II (IGF-I... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2cnj | ||||||
|---|---|---|---|---|---|---|---|
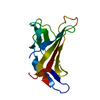
| Title | NMR studies on the interaction of Insulin-Growth Factor II (IGF-II) with IGF2R domain 11 | ||||||
 Components Components | CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR | ||||||
 Keywords Keywords | RECEPTOR / TRANSPORT / POLYMORPHISM / GLYCOPROTEIN / TRANSMEMBRANE / IGF2R / CANCER / MEMBRANE / LYSOSOME / INSULIN-LIKE GROWTH FACTOR-II | ||||||
| Function / homology |  Function and homology information Function and homology informationclathrin coat / Retrograde transport at the Trans-Golgi-Network / response to tetrachloromethane / insulin-like growth factor receptor activity / retromer complex binding / insulin-like growth factor binding / insulin-like growth factor II binding / trans-Golgi network transport vesicle / host-mediated activation of viral process / retinoic acid binding ...clathrin coat / Retrograde transport at the Trans-Golgi-Network / response to tetrachloromethane / insulin-like growth factor receptor activity / retromer complex binding / insulin-like growth factor binding / insulin-like growth factor II binding / trans-Golgi network transport vesicle / host-mediated activation of viral process / retinoic acid binding / lysosomal transport / Golgi Associated Vesicle Biogenesis / nuclear envelope lumen / D-mannose binding / animal organ regeneration / G-protein alpha-subunit binding / endocytic vesicle / response to retinoic acid / transport vesicle / receptor-mediated endocytosis / secretory granule membrane / trans-Golgi network membrane / post-embryonic development / phosphoprotein binding / trans-Golgi network / clathrin-coated endocytic vesicle membrane / liver development / late endosome / Cargo recognition for clathrin-mediated endocytosis / signaling receptor activity / Clathrin-mediated endocytosis / spermatogenesis / early endosome / endosome / endosome membrane / positive regulation of apoptotic process / G protein-coupled receptor signaling pathway / Golgi membrane / focal adhesion / Neutrophil degranulation / perinuclear region of cytoplasm / enzyme binding / cell surface / Golgi apparatus / signal transduction / extracellular exosome / identical protein binding / membrane / plasma membrane Similarity search - Function | ||||||
| Biological species |  HOMO SAPIENS (human) HOMO SAPIENS (human) | ||||||
| Method | SOLUTION NMR / WATER REFINED IN CNS | ||||||
 Authors Authors | Williams, C. / Prince, S. / Zaccheo, O. / Hassan, A.B. / Crosby, J. / Crump, M. | ||||||
 Citation Citation |  Journal: Structure / Year: 2007 Journal: Structure / Year: 2007Title: Structural Insights Into the Interaction of Insulin-Like Growth Factor 2 with Igf2R Domain 11. Authors: Williams, C. / Rezgui, D. / Prince, S.N. / Zaccheo, O.J. / Foulstone, E.J. / Forbes, B.E. / Norton, R.S. / Crosby, J. / Hassan, A.B. / Crump, M.P. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2cnj.cif.gz 2cnj.cif.gz | 839.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2cnj.ent.gz pdb2cnj.ent.gz | 700.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2cnj.json.gz 2cnj.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/cn/2cnj https://data.pdbj.org/pub/pdb/validation_reports/cn/2cnj ftp://data.pdbj.org/pub/pdb/validation_reports/cn/2cnj ftp://data.pdbj.org/pub/pdb/validation_reports/cn/2cnj | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 16651.848 Da / Num. of mol.: 1 / Fragment: DOMAIN 11, RESIDUES 1508-1650 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  HOMO SAPIENS (human) / Plasmid: PET22 / Production host: HOMO SAPIENS (human) / Plasmid: PET22 / Production host:  |
|---|---|
| Compound details | PROMOTES THE TRANSPORT OF PHOSPHORYL |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||
| NMR details | Text: THE STRUCTURE WAS DETERMINED USING TRIPLE-RESONANCE NMR SPECTROSCOPY ON UNLABELLED, 15N AND 13C, 15N-LABELED DOMAIN 11. |
- Sample preparation
Sample preparation
| Details | Contents: 5% D2O/ 95% H2O |
|---|---|
| Sample conditions | pH: 5.5 / Pressure: 1.0 atm / Temperature: 298.0 K |
-NMR measurement
| NMR spectrometer | Type: Varian INOVA / Manufacturer: Varian / Model: INOVA / Field strength: 600 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: WATER REFINED IN CNS / Software ordinal: 1 Details: WATER REFINED USING RECORRD SCIPTS IN CNS. A.J.NEDERVEEN,J.F.DORELEIJERS,W.VRANKEN, Z.MILLER, C.A.SPRONK, S.B.NABUURS, P.GUNTERT, M.LIVNY, J.L.MARKLEY, M.NILGES, E.L.ULRICH, R.KAPTEIN AND A. ...Details: WATER REFINED USING RECORRD SCIPTS IN CNS. A.J.NEDERVEEN,J.F.DORELEIJERS,W.VRANKEN, Z.MILLER, C.A.SPRONK, S.B.NABUURS, P.GUNTERT, M.LIVNY, J.L.MARKLEY, M.NILGES, E.L.ULRICH, R.KAPTEIN AND A.M.BONVIN, PROTEINS. RECOORD: A RECALCULATED COORDINATES DATABASE OF 500+ PROTEINS FROM THE PDB USING RESTRAINTS FROM THE BIOMAGRESBANK. PROTEINS 59, 662-672. | ||||||||||||||||||
| NMR ensemble | Conformer selection criteria: LOWEST ENERGY, LEAST VIOLATION Conformers calculated total number: 200 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller















 PDBj
PDBj



 HSQC
HSQC