+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2b6g | ||||||
|---|---|---|---|---|---|---|---|
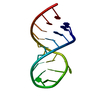
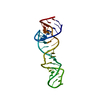
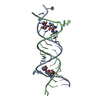
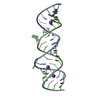
| Title | RNA recognition by the Vts1 SAM domain | ||||||
 Components Components |
| ||||||
 Keywords Keywords | RNA BINDING PROTEIN / alpha-helix / pentaloop / hairpin | ||||||
| Function / homology |  Function and homology information Function and homology informationflap-structured DNA binding / positive regulation of DNA metabolic process / nuclear-transcribed mRNA poly(A) tail shortening / nuclear-transcribed mRNA catabolic process / P-body / protein transport / nucleotide binding / mRNA binding / RNA binding / nucleus / cytosol Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method | SOLUTION NMR / simulated annealing | ||||||
 Authors Authors | Donaldson, L.W. / Johnson, P.E. | ||||||
 Citation Citation |  Journal: Nat.Struct.Mol.Biol. / Year: 2006 Journal: Nat.Struct.Mol.Biol. / Year: 2006Title: RNA recognition by the Vts1p SAM domain Authors: Johnson, P.E. / Donaldson, L.W. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2b6g.cif.gz 2b6g.cif.gz | 53.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2b6g.ent.gz pdb2b6g.ent.gz | 36.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2b6g.json.gz 2b6g.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  2b6g_validation.pdf.gz 2b6g_validation.pdf.gz | 304.7 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  2b6g_full_validation.pdf.gz 2b6g_full_validation.pdf.gz | 304.6 KB | Display | |
| Data in XML |  2b6g_validation.xml.gz 2b6g_validation.xml.gz | 3.7 KB | Display | |
| Data in CIF |  2b6g_validation.cif.gz 2b6g_validation.cif.gz | 4.9 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/b6/2b6g https://data.pdbj.org/pub/pdb/validation_reports/b6/2b6g ftp://data.pdbj.org/pub/pdb/validation_reports/b6/2b6g ftp://data.pdbj.org/pub/pdb/validation_reports/b6/2b6g | HTTPS FTP |
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 6086.633 Da / Num. of mol.: 1 / Fragment: Smaug Recognition Element / Source method: obtained synthetically Details: RNA was produced by T7 polymerase based in vitro transcription |
|---|---|
| #2: Protein | Mass: 13096.857 Da / Num. of mol.: 1 / Fragment: SAM domain Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Gene: Vts1p / Plasmid: pGEX / Species (production host): Escherichia coli / Production host:  |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Contents: 0.2-0.8 mM U-15N,13C; 20 mM sodium phosphate buffer, pH 7.8, 150 mM sodium chloride, 0.02 % sodium azide Solvent system: 90% H2O/10% D2O |
|---|---|
| Sample conditions | Ionic strength: 150 mM NaCl / pH: 7.8 / Pressure: ambient / Temperature: 293 K |
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Radiation wavelength | Relative weight: 1 | ||||||||||||||||||||
| NMR spectrometer |
|
- Processing
Processing
| NMR software |
| ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing / Software ordinal: 1 Details: Protein NOE restraints were calibrated from peak volumes to distances ranging from 1.8-5.0 using the CANDID module of CYANA v2.1. Initially, 100 structures were calculated with XPLOR-NIH v2. ...Details: Protein NOE restraints were calibrated from peak volumes to distances ranging from 1.8-5.0 using the CANDID module of CYANA v2.1. Initially, 100 structures were calculated with XPLOR-NIH v2.11.0 starting from a partially docked protein-RNA complex. A simulated annealing approach with internal torsion angle dynamics, delphic database potentials and RNA planarity restraints was used. From the initial ensemble of structures, 20 were selected based upon low energy, no NOE violations > 0.5 angstroms and no dihedral angle violations > 5 degrees. Restraints used for the structure calculation: 882 intraresidue protein-protein NOE, 435 sequential protein-protein NOE, 335 medium range protein-protein NOE, 263 long range protein-protein NOE, 80 protein hydrogen bonds, 60 RNA-RNA NOE, 23 protein-RNA NOE, 140 protein dihedral angle, 93 RNA dihedral angle. | ||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller











 PDBj
PDBj





























 CYANA
CYANA