| Software | | Name | Version | Classification |
|---|
| ADSC | Quantumdata collection| PHENIX | | model building| CNS | 1.3 | refinement| CrystalClear | | data reduction| CrystalClear | | data scaling| PHENIX | | phasing | | | | | | |
|
|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 2.8→30 Å / Occupancy max: 1 / Occupancy min: 1 MOLECULAR REPLACEMENT / Resolution: 2.8→30 Å / Occupancy max: 1 / Occupancy min: 1
| Rfactor | Num. reflection | % reflection |
|---|
| Rfree | 0.227 | 330 | 9 % |
|---|
| Rwork | 0.201 | - | - |
|---|
| obs | - | 3344 | 91.1 % |
|---|
|
|---|
| Solvent computation | Bsol: 34.3561 Å2 |
|---|
| Displacement parameters | Biso max: 103.49 Å2 / Biso mean: 43.0193 Å2 / Biso min: 9.55 Å2
| Baniso -1 | Baniso -2 | Baniso -3 |
|---|
| 1- | -1.616 Å2 | 0 Å2 | 0 Å2 |
|---|
| 2- | - | 15.862 Å2 | 0 Å2 |
|---|
| 3- | - | - | -14.246 Å2 |
|---|
|
|---|
| Refinement step | Cycle: LAST / Resolution: 2.8→30 Å
| Protein | Nucleic acid | Ligand | Solvent | Total |
|---|
| Num. atoms | 0 | 940 | 62 | 36 | 1038 |
|---|
|
|---|
| Refine LS restraints | | Refine-ID | Type | Dev ideal | Dev ideal target |
|---|
| X-RAY DIFFRACTION | c_bond_d| 0.005 | | | X-RAY DIFFRACTION | c_angle_d| 0.969 | | | X-RAY DIFFRACTION | c_mcbond_it| 0 | 1.5 | | X-RAY DIFFRACTION | c_scbond_it| 2.467 | 2 | | X-RAY DIFFRACTION | c_mcangle_it| 0 | 2 | | X-RAY DIFFRACTION | c_scangle_it| 3.659 | 2.5 | | | | | | |
|
|---|
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 10 | Resolution (Å) | Rfactor Rfree | Num. reflection Rfree | Rfactor Rwork | Num. reflection Rwork | Num. reflection all | % reflection obs (%) |
|---|
| 2.8-2.9 | 0.3356 | 26 | 0.3827 | 301 | 327 | 93.7 | | 2.9-3.02 | 0.4144 | 34 | 0.3258 | 295 | 329 | 92.2 | | 3.02-3.15 | 0.2856 | 44 | 0.284 | 286 | 330 | 92.2 | | 3.15-3.32 | 0.2458 | 31 | 0.2576 | 283 | 314 | 87.5 | | 3.32-3.53 | 0.3498 | 33 | 0.2398 | 281 | 314 | 88.2 | | 3.53-3.8 | 0.1909 | 41 | 0.1807 | 283 | 324 | 88.3 | | 3.8-4.18 | 0.2155 | 36 | 0.1985 | 283 | 319 | 87.4 | | 4.18-4.78 | 0.1743 | 30 | 0.1498 | 303 | 333 | 89.3 | | 4.78-6.02 | 0.115 | 21 | 0.1308 | 337 | 358 | 95.2 | | 6.02-30 | 0.191 | 34 | 0.168 | 362 | 396 | 96.4 |
|
|---|
| Xplor file | | Refine-ID | Serial no | Param file |
|---|
| X-RAY DIFFRACTION | 1 | dna-rna_free.param | | X-RAY DIFFRACTION | 2 | water_rep.param| X-RAY DIFFRACTION | 3 | ion.param| X-RAY DIFFRACTION | 4 | 115_xplor.param | | |
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
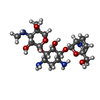
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.8 Å
MOLECULAR REPLACEMENT / Resolution: 2.8 Å  Authors
Authors Citation
Citation Journal: Chemmedchem / Year: 2013
Journal: Chemmedchem / Year: 2013 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 4gpy.cif.gz
4gpy.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb4gpy.ent.gz
pdb4gpy.ent.gz PDB format
PDB format 4gpy.json.gz
4gpy.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/gp/4gpy
https://data.pdbj.org/pub/pdb/validation_reports/gp/4gpy ftp://data.pdbj.org/pub/pdb/validation_reports/gp/4gpy
ftp://data.pdbj.org/pub/pdb/validation_reports/gp/4gpy Links
Links Assembly
Assembly
 Components
Components X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  Photon Factory
Photon Factory  / Beamline: AR-NW12A / Wavelength: 1 Å
/ Beamline: AR-NW12A / Wavelength: 1 Å Processing
Processing MOLECULAR REPLACEMENT / Resolution: 2.8→30 Å / Occupancy max: 1 / Occupancy min: 1
MOLECULAR REPLACEMENT / Resolution: 2.8→30 Å / Occupancy max: 1 / Occupancy min: 1  Movie
Movie Controller
Controller








 PDBj
PDBj