+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: PDB / ID: 1vbe | ||||||
|---|---|---|---|---|---|---|---|
| タイトル | POLIOVIRUS (TYPE 3, SABIN STRAIN, MUTANT 242-H2) COMPLEXED WITH R78206 | ||||||
 要素 要素 | (POLIOVIRUS TYPE ...) x 5 | ||||||
 キーワード キーワード | VIRUS / VIRUS COAT PROTEIN / HYDROLASE / THIOL PROTEASE / Icosahedral virus | ||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報caveolin-mediated endocytosis of virus by host cell / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MDA-5 activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / ribonucleoside triphosphate phosphatase activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid ...caveolin-mediated endocytosis of virus by host cell / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MDA-5 activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / ribonucleoside triphosphate phosphatase activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / nucleoside-triphosphate phosphatase / channel activity / monoatomic ion transmembrane transport / DNA replication / RNA helicase activity / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / virion attachment to host cell / host cell nucleus / structural molecule activity / proteolysis / RNA binding / zinc ion binding / ATP binding / membrane 類似検索 - 分子機能 | ||||||
| 生物種 | Poliovirus type 3 | ||||||
| 手法 |  X線回折 / 解像度: 2.8 Å X線回折 / 解像度: 2.8 Å | ||||||
 データ登録者 データ登録者 | Grant, R.A. / Hiremath, C.N. / Filman, D.J. / Syed, R. / Andries, K. / Hogle, J.M. | ||||||
 引用 引用 |  ジャーナル: Curr.Biol. / 年: 1994 ジャーナル: Curr.Biol. / 年: 1994タイトル: Structures of poliovirus complexes with anti-viral drugs: implications for viral stability and drug design. 著者: Grant, R.A. / Hiremath, C.N. / Filman, D.J. / Syed, R. / Andries, K. / Hogle, J.M. #1:  ジャーナル: Acta Crystallogr.,Sect.D / 年: 1995 ジャーナル: Acta Crystallogr.,Sect.D / 年: 1995タイトル: Binding of the Antiviral Drug Win51711 to the Sabin Strain of Type 3 Poliovirus: Structural Comparison with Drug Binding in Rhinovirus 14 著者: Hiremath, C.N. / Grant, R.A. / Filman, D.J. / Hogle, J.M. #2:  ジャーナル: New Aspects of Positive-Strand RNA Viruses / 年: 1990 ジャーナル: New Aspects of Positive-Strand RNA Viruses / 年: 1990タイトル: Role of Conformational Transitions in Poliovirus Assembly and Cell Entry 著者: Hogle, J.M. / Syed, R. / Fricks, C.E. / Icenogle, J.P. / Flore, O. / Filman, D.J. #3:  ジャーナル: Embo J. / 年: 1989 ジャーナル: Embo J. / 年: 1989タイトル: Structural Factors that Control Conformational Transitions and Serotype Specificity in Type 3 Poliovirus 著者: Filman, D.J. / Syed, R. / Chow, M. / Macadam, A.J. / Minor, P.D. / Hogle, J.M. #4:  ジャーナル: Nature / 年: 1987 ジャーナル: Nature / 年: 1987タイトル: Myristylation of Picornavirus Capsid Protein Vp4 and its Structural Significance 著者: Chow, M. / Newman, J.F. / Filman, D. / Hogle, J.M. / Rowlands, D.J. / Brown, F. #5:  ジャーナル: Science / 年: 1985 ジャーナル: Science / 年: 1985タイトル: Three-Dimensional Structure of Poliovirus at 2.9 A Resolution 著者: Hogle, J.M. / Chow, M. / Filman, D.J. #6:  ジャーナル: Nucleic Acids Res. / 年: 1983 ジャーナル: Nucleic Acids Res. / 年: 1983タイトル: The Nucleotide Sequence of Poliovirus Type 3 Leon 12 A1B: Comparison with Poliovirus Type 1 著者: Stanway, G. / Cann, A.J. / Hauptmann, R. / Hughes, P. / Clarke, L.D. / Mountford, R.C. / Minor, P.D. / Schild, G.C. / Almond, J.W. | ||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 構造ビューア | 分子:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- ダウンロードとリンク
ダウンロードとリンク
- ダウンロード
ダウンロード
| PDBx/mmCIF形式 |  1vbe.cif.gz 1vbe.cif.gz | 179.7 KB | 表示 |  PDBx/mmCIF形式 PDBx/mmCIF形式 |
|---|---|---|---|---|
| PDB形式 |  pdb1vbe.ent.gz pdb1vbe.ent.gz | 140.8 KB | 表示 |  PDB形式 PDB形式 |
| PDBx/mmJSON形式 |  1vbe.json.gz 1vbe.json.gz | ツリー表示 |  PDBx/mmJSON形式 PDBx/mmJSON形式 | |
| その他 |  その他のダウンロード その他のダウンロード |
-検証レポート
| 文書・要旨 |  1vbe_validation.pdf.gz 1vbe_validation.pdf.gz | 476.3 KB | 表示 |  wwPDB検証レポート wwPDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  1vbe_full_validation.pdf.gz 1vbe_full_validation.pdf.gz | 487.9 KB | 表示 | |
| XML形式データ |  1vbe_validation.xml.gz 1vbe_validation.xml.gz | 18.8 KB | 表示 | |
| CIF形式データ |  1vbe_validation.cif.gz 1vbe_validation.cif.gz | 28.5 KB | 表示 | |
| アーカイブディレクトリ |  https://data.pdbj.org/pub/pdb/validation_reports/vb/1vbe https://data.pdbj.org/pub/pdb/validation_reports/vb/1vbe ftp://data.pdbj.org/pub/pdb/validation_reports/vb/1vbe ftp://data.pdbj.org/pub/pdb/validation_reports/vb/1vbe | HTTPS FTP |
-関連構造データ
- リンク
リンク
- 集合体
集合体
| 登録構造単位 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | x 60
| ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
| ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | x 5
| ||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | x 6
| ||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | x 15
| ||||||||||||||||||||||||||||||||||||||||||||||||
| 単位格子 |
| ||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 点対称性: (ヘルマン・モーガン記号: 532 / シェーンフリース記号: I (正20面体型対称)) | ||||||||||||||||||||||||||||||||||||||||||||||||
| 非結晶学的対称性 (NCS) | NCS oper:
|
- 要素
要素
-POLIOVIRUS TYPE ... , 5種, 5分子 01234
| #1: タンパク質・ペプチド | 分子量: 446.495 Da / 分子数: 1 / 変異: CHAIN 1, F124L, F134L / 由来タイプ: 天然 詳細: P3/242-H2 DERIVED FROM A LOW-PASSAGE SEED STOCK OF A PLAQUE ISOLATE PROVIDED BY A. MACADAM (NATIONAL INSTITUTE FOR BIOLOGICAL STANDARDS CONTROL, LONDON) 由来: (天然)  Poliovirus type 3 (strains P3/LEON/37 AND P3/LEON 12A[1]B) (ポリオウイルス) Poliovirus type 3 (strains P3/LEON/37 AND P3/LEON 12A[1]B) (ポリオウイルス)属: Enterovirus / 生物種: Poliovirus / 株: P3-242-H2 |
|---|---|
| #2: タンパク質 | 分子量: 33455.641 Da / 分子数: 1 / 変異: CHAIN 1, F124L, F134L / 由来タイプ: 天然 詳細: P3/242-H2 DERIVED FROM A LOW-PASSAGE SEED STOCK OF A PLAQUE ISOLATE PROVIDED BY A. MACADAM (NATIONAL INSTITUTE FOR BIOLOGICAL STANDARDS CONTROL, LONDON) 由来: (天然)  Poliovirus type 3 (strains P3/LEON/37 AND P3/LEON 12A[1]B) (ポリオウイルス) Poliovirus type 3 (strains P3/LEON/37 AND P3/LEON 12A[1]B) (ポリオウイルス)属: Enterovirus / 器官: SEED / 生物種: Poliovirus / 株: P3-242-H2 / 参照: UniProt: P03302 |
| #3: タンパク質 | 分子量: 30169.920 Da / 分子数: 1 / 変異: CHAIN 1, F124L, F134L / 由来タイプ: 天然 詳細: P3/242-H2 DERIVED FROM A LOW-PASSAGE SEED STOCK OF A PLAQUE ISOLATE PROVIDED BY A. MACADAM (NATIONAL INSTITUTE FOR BIOLOGICAL STANDARDS CONTROL, LONDON) 由来: (天然)  Poliovirus type 3 (strains P3/LEON/37 AND P3/LEON 12A[1]B) (ポリオウイルス) Poliovirus type 3 (strains P3/LEON/37 AND P3/LEON 12A[1]B) (ポリオウイルス)属: Enterovirus / 器官: SEED / 生物種: Poliovirus / 株: P3-242-H2 / 参照: UniProt: P03302 |
| #4: タンパク質 | 分子量: 25950.723 Da / 分子数: 1 / 変異: CHAIN 1, F124L, F134L / 由来タイプ: 天然 詳細: P3/242-H2 DERIVED FROM A LOW-PASSAGE SEED STOCK OF A PLAQUE ISOLATE PROVIDED BY A. MACADAM (NATIONAL INSTITUTE FOR BIOLOGICAL STANDARDS CONTROL, LONDON) 由来: (天然)  Poliovirus type 3 (strains P3/LEON/37 AND P3/LEON 12A[1]B) (ポリオウイルス) Poliovirus type 3 (strains P3/LEON/37 AND P3/LEON 12A[1]B) (ポリオウイルス)属: Enterovirus / 器官: SEED / 生物種: Poliovirus / 株: P3-242-H2 / 参照: UniProt: P03302 |
| #5: タンパク質 | 分子量: 7320.917 Da / 分子数: 1 / 変異: CHAIN 1, F124L, F134L / 由来タイプ: 天然 詳細: P3/242-H2 DERIVED FROM A LOW-PASSAGE SEED STOCK OF A PLAQUE ISOLATE PROVIDED BY A. MACADAM (NATIONAL INSTITUTE FOR BIOLOGICAL STANDARDS CONTROL, LONDON) 由来: (天然)  Poliovirus type 3 (strains P3/LEON/37 AND P3/LEON 12A[1]B) (ポリオウイルス) Poliovirus type 3 (strains P3/LEON/37 AND P3/LEON 12A[1]B) (ポリオウイルス)属: Enterovirus / 器官: SEED / 生物種: Poliovirus / 株: P3-242-H2 / 参照: UniProt: P03302 |
-非ポリマー , 2種, 2分子 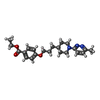


| #6: 化合物 | ChemComp-J78 / ( |
|---|---|
| #7: 化合物 | ChemComp-MYR / |
-詳細
| Has protein modification | Y |
|---|---|
| 配列の詳細 | THE APPROPRIATE SEQUENCE FOR THIS VIRUS CORRESPONDS TO THE STANWAY ET AL. REFERENCE ABOVE EXCEPT ...THE APPROPRIAT |
-実験情報
-実験
| 実験 | 手法:  X線回折 X線回折 |
|---|
- 試料調製
試料調製
| 結晶 | マシュー密度: 2.59 Å3/Da / 溶媒含有率: 30 % |
|---|---|
| 結晶化 | *PLUS 手法: other / 詳細: Filman, D.J., (1989) EMBO, J., 8, 1567. |
-データ収集
| 放射 | 散乱光タイプ: x-ray |
|---|---|
| 放射波長 | 相対比: 1 |
| 反射 | 解像度: 2.8→30 Å / Num. obs: 161423 / % possible obs: 30 % / Observed criterion σ(I): 0 |
| 反射 | *PLUS Num. measured all: 349922 |
- 解析
解析
| ソフトウェア |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 精密化 | Rfactor Rwork: 0.295 / 最高解像度: 2.8 Å 詳細: STEREOCHEMICAL CONSTRAINTS BASED ON PARAM19 AND TOP19 FILES USED IN X-PLOR VERSIONS PREVIOUS TO VERSION 3.1 ATOMIC MODELS FOR THE VIRUS AND THE DRUG WERE BUILT TO FIT ICOSAHEDRALLY ...詳細: STEREOCHEMICAL CONSTRAINTS BASED ON PARAM19 AND TOP19 FILES USED IN X-PLOR VERSIONS PREVIOUS TO VERSION 3.1 ATOMIC MODELS FOR THE VIRUS AND THE DRUG WERE BUILT TO FIT ICOSAHEDRALLY CONSTRAINED 'FO' MAP USING THE GRAPHICS PROGRAM FRODO (JONES, 1978) MODIFIED TO INCORPORATE T = 1 ICOSAHEDRAL SYMMETRY. ATOMIC MODELS WERE OPTIMIZED WITH RESPECT TO THIS MAP BY A PSEUDO-REAL-SPACE REFINEMENT PROCEDURE, MINIMIZING A RESIDUAL WITH A STEREOCHEMICAL AND A CRYSTALLOGRAPHIC COMPONENT. THE GRADIENT OF THE STEREOCHEMICAL COMPONENT WAS PROVIDED BY THE X-PLOR PROGRAM (A. BRUNGER, X-PLOR VERSION 2.1 YALE UNIVERSITY 1990). THE CRYSTALLOGRAPHIC COMPONENT AND ITS GRADIENT WERE EVALUATED OVER THE VOLUME OF AN ARBITRARY PSEUDO-CELL (THE PROTOMER BOX) WHICH IS SUFFICIENTLY LARGE TO COMFORTABLY ENCLOSE A COMPLETE CHEMICALLY CONTINUOUS POLIOVIRUS PROTOMER, TOGETHER WITH WHATEVER FRAGMENTS OF SYMMETRY-RELATED PROTOMERS HAPPEN TO LIE SUFFICIENTLY CLOSE TO THE BOX TO CONTRIBUTE TO IT. THIS REFINEMENT SEEKS TO MINIMIZE THE DISCREPANCY BETWEEN THE 'PHASED' FOURIER TRANSFORMS OF MODEL-BASED ELECTRON DENSITY AND AUTHENTIC SYMMETRY-CONSTRAINED ELECTRON DENSITY, WHEN EACH TRANSFORM IS CALCULATED IN THE ARBITRARY PROTOMER BOX VOLUME, SCALED IN A RESOLUTION-DEPENDENT FASHION. SEE JRNL REFERENCE FOR MORE DETAILS. THE DISORDERED RESIDUES ABSENT FROM THE MODEL INCLUDE THE AMINO TERMINUS OF VP1 PRIOR TO THE RESIDUE LABELED GLN 24, 1 - 5 IN VP2, 236 - 238 IN VP3, AND THE RESIDUES LABELED 17 - 22 IN VP4. THE POLYPEPTIDE DESIGNATED IN THIS FILE AS RESIDUES 6 - 9 OF CHAIN 0 REPRESENTS A FEATURE IN THE ELECTRON DENSITY MAP WHICH APPEARS TO BE A BETA STRAND. ALTHOUGH THE SIDE CHAINS OF THIS STRAND CANNOT BE CORRELATED RELIABLY WITH THE SEQUENCE OF THE PROTEIN, THE FEATURE IS BELIEVED LIKELY TO CORRESPOND TO SOME PORTION OF THE AMINO TERMINAL EXTENSION OF VP1. NO SOLVENT MOLECULES ARE INCLUDED. THE POLYPEPTIDE DESIGNATED IN THIS FILE AS RESIDUES 6 - 9 OF CHAIN 0 REPRESENTS A FEATURE IN THE ELECTRON DENSITY MAP WHICH APPEARS TO BE A BETA STRAND. ALTHOUGH THE SIDE CHAINS OF THIS STRAND CANNOT BE CORRELATED RELIABLY WITH THE SEQUENCE OF THE PROTEIN, THE FEATURE IS BELIEVED LIKELY TO CORRESPOND TO SOME PORTION OF THE AMINO TERMINAL EXTENSION OF VP1. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 精密化ステップ | サイクル: LAST / 最高解像度: 2.8 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 拘束条件 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ソフトウェア | *PLUS 名称: 'PSEUDO-REAL SPACE PROCEDURE' / 分類: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 精密化 | *PLUS Rfactor obs: 0.295 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 溶媒の処理 | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 原子変位パラメータ | *PLUS |
 ムービー
ムービー コントローラー
コントローラー



























 PDBj
PDBj






