[English] 日本語
 Yorodumi
Yorodumi- PDB-1vba: POLIOVIRUS (TYPE 3, SABIN STRAIN) (P3/SABIN, P3/LEON/12A(1)B) COM... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1vba | ||||||
|---|---|---|---|---|---|---|---|
| Title | POLIOVIRUS (TYPE 3, SABIN STRAIN) (P3/SABIN, P3/LEON/12A(1)B) COMPLEXED WITH R78206 | ||||||
 Components Components | (POLIOVIRUS TYPE ...) x 5 | ||||||
 Keywords Keywords | VIRUS / VIRUS COAT PROTEIN / HYDROLASE / THIOL PROTEASE / Icosahedral virus | ||||||
| Function / homology |  Function and homology information Function and homology informationcaveolin-mediated endocytosis of virus by host cell / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MDA-5 activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / ribonucleoside triphosphate phosphatase activity ...caveolin-mediated endocytosis of virus by host cell / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MDA-5 activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / ribonucleoside triphosphate phosphatase activity / host cell cytoplasmic vesicle membrane / nucleoside-triphosphate phosphatase / channel activity / monoatomic ion transmembrane transport / DNA replication / RNA helicase activity / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / virion attachment to host cell / host cell nucleus / structural molecule activity / proteolysis / RNA binding / zinc ion binding / ATP binding / membrane Similarity search - Function | ||||||
| Biological species | Poliovirus type 3 | ||||||
| Method |  X-RAY DIFFRACTION / Resolution: 2.9 Å X-RAY DIFFRACTION / Resolution: 2.9 Å | ||||||
 Authors Authors | Grant, R.A. / Hiremath, C.N. / Filman, D.J. / Syed, R. / Andries, K. / Hogle, J.M. | ||||||
 Citation Citation |  Journal: Curr.Biol. / Year: 1994 Journal: Curr.Biol. / Year: 1994Title: Structures of poliovirus complexes with anti-viral drugs: implications for viral stability and drug design. Authors: Grant, R.A. / Hiremath, C.N. / Filman, D.J. / Syed, R. / Andries, K. / Hogle, J.M. #1:  Journal: Acta Crystallogr.,Sect.D / Year: 1995 Journal: Acta Crystallogr.,Sect.D / Year: 1995Title: Binding of the Antiviral Drug Win51711 to the Sabin Strain of Type 3 Poliovirus: Structural Comparison with Drug Binding in Rhinovirus 14 Authors: Hiremath, C.N. / Grant, R.A. / Filman, D.J. / Hogle, J.M. #2:  Journal: New Aspects of Positive-Strand RNA Viruses / Year: 1990 Journal: New Aspects of Positive-Strand RNA Viruses / Year: 1990Title: Role of Conformational Transitions in Poliovirus Assembly and Cell Entry Authors: Hogle, J.M. / Syed, R. / Fricks, C.E. / Icenogle, J.P. / Flore, O. / Filman, D.J. #3:  Journal: Embo J. / Year: 1989 Journal: Embo J. / Year: 1989Title: Structural Factors that Control Conformational Transitions and Serotype Specificity in Type 3 Poliovirus Authors: Filman, D.J. / Syed, R. / Chow, M. / Macadam, A.J. / Minor, P.D. / Hogle, J.M. #4:  Journal: Nature / Year: 1987 Journal: Nature / Year: 1987Title: Myristylation of Picornavirus Capsid Protein Vp4 and its Structural Significance Authors: Chow, M. / Newman, J.F. / Filman, D. / Hogle, J.M. / Rowlands, D.J. / Brown, F. #5:  Journal: Science / Year: 1985 Journal: Science / Year: 1985Title: Three-Dimensional Structure of Poliovirus at 2.9 A Resolution Authors: Hogle, J.M. / Chow, M. / Filman, D.J. #6:  Journal: Nucleic Acids Res. / Year: 1983 Journal: Nucleic Acids Res. / Year: 1983Title: The Nucleotide Sequence of Poliovirus Type 3 Leon 12 A1B: Comparison with Poliovirus Type 1 Authors: Stanway, G. / Cann, A.J. / Hauptmann, R. / Hughes, P. / Clarke, L.D. / Mountford, R.C. / Minor, P.D. / Schild, G.C. / Almond, J.W. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1vba.cif.gz 1vba.cif.gz | 180.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1vba.ent.gz pdb1vba.ent.gz | 142.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1vba.json.gz 1vba.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/vb/1vba https://data.pdbj.org/pub/pdb/validation_reports/vb/1vba ftp://data.pdbj.org/pub/pdb/validation_reports/vb/1vba ftp://data.pdbj.org/pub/pdb/validation_reports/vb/1vba | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | x 60
| ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
| ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | x 5
| ||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | x 6
| ||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | x 15
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Unit cell |
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: (Hermann–Mauguin notation: 532 / Schoenflies symbol: I (icosahedral)) | ||||||||||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS oper:
|
- Components
Components
-POLIOVIRUS TYPE ... , 5 types, 5 molecules 01234
| #1: Protein/peptide | Mass: 446.495 Da / Num. of mol.: 1 / Source method: isolated from a natural source Details: P3/SABIN PREPARED FROM A LOW-PASSAGE SEED STOCK OF A PLAQUE ISOLATE (P3/LEON/12A(1)B PLACQUE 411) OBTAINED FROM P.D.MINOR (NATIONAL INSTITUTE FOR BIOLOGICAL STANDARDS CONTROL, LONDON) Source: (natural)  Poliovirus type 3 (strains P3/LEON/37 AND P3/LEON 12A[1]B) Poliovirus type 3 (strains P3/LEON/37 AND P3/LEON 12A[1]B)Genus: Enterovirus / Species: Poliovirus / Strain: P3-SABIN |
|---|---|
| #2: Protein | Mass: 33523.672 Da / Num. of mol.: 1 / Source method: isolated from a natural source Details: P3/SABIN PREPARED FROM A LOW-PASSAGE SEED STOCK OF A PLAQUE ISOLATE (P3/LEON/12A(1)B PLACQUE 411) OBTAINED FROM P.D.MINOR (NATIONAL INSTITUTE FOR BIOLOGICAL STANDARDS CONTROL, LONDON) Source: (natural)  Poliovirus type 3 (strains P3/LEON/37 AND P3/LEON 12A[1]B) Poliovirus type 3 (strains P3/LEON/37 AND P3/LEON 12A[1]B)Genus: Enterovirus / Organ: SEED / Species: Poliovirus / Strain: P3-SABIN / References: UniProt: P03302 |
| #3: Protein | Mass: 30169.920 Da / Num. of mol.: 1 / Source method: isolated from a natural source Details: P3/SABIN PREPARED FROM A LOW-PASSAGE SEED STOCK OF A PLAQUE ISOLATE (P3/LEON/12A(1)B PLACQUE 411) OBTAINED FROM P.D.MINOR (NATIONAL INSTITUTE FOR BIOLOGICAL STANDARDS CONTROL, LONDON) Source: (natural)  Poliovirus type 3 (strains P3/LEON/37 AND P3/LEON 12A[1]B) Poliovirus type 3 (strains P3/LEON/37 AND P3/LEON 12A[1]B)Genus: Enterovirus / Organ: SEED / Species: Poliovirus / Strain: P3-SABIN / References: UniProt: P03302 |
| #4: Protein | Mass: 25950.723 Da / Num. of mol.: 1 / Source method: isolated from a natural source Details: P3/SABIN PREPARED FROM A LOW-PASSAGE SEED STOCK OF A PLAQUE ISOLATE (P3/LEON/12A(1)B PLACQUE 411) OBTAINED FROM P.D.MINOR (NATIONAL INSTITUTE FOR BIOLOGICAL STANDARDS CONTROL, LONDON) Source: (natural)  Poliovirus type 3 (strains P3/LEON/37 AND P3/LEON 12A[1]B) Poliovirus type 3 (strains P3/LEON/37 AND P3/LEON 12A[1]B)Genus: Enterovirus / Organ: SEED / Species: Poliovirus / Strain: P3-SABIN / References: UniProt: P03302 |
| #5: Protein | Mass: 7320.917 Da / Num. of mol.: 1 / Source method: isolated from a natural source Details: P3/SABIN PREPARED FROM A LOW-PASSAGE SEED STOCK OF A PLAQUE ISOLATE (P3/LEON/12A(1)B PLACQUE 411) OBTAINED FROM P.D.MINOR (NATIONAL INSTITUTE FOR BIOLOGICAL STANDARDS CONTROL, LONDON) Source: (natural)  Poliovirus type 3 (strains P3/LEON/37 AND P3/LEON 12A[1]B) Poliovirus type 3 (strains P3/LEON/37 AND P3/LEON 12A[1]B)Genus: Enterovirus / Organ: SEED / Species: Poliovirus / Strain: P3-SABIN / References: UniProt: P03302 |
-Non-polymers , 2 types, 2 molecules 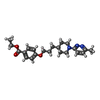


| #6: Chemical | ChemComp-J78 / ( |
|---|---|
| #7: Chemical | ChemComp-MYR / |
-Details
| Has protein modification | Y |
|---|---|
| Sequence details | THE APPROPRIATE SEQUENCE FOR THIS VIRUS CORRESPONDS TO THE STANWAY ET AL. REFERENCE ABOVE. THE ...THE APPROPRIAT |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.59 Å3/Da / Density % sol: 30 % |
|---|---|
| Crystal grow | *PLUS Method: other / Details: Filman, D.J., (1989) EMBO, J., 8, 1567. |
-Data collection
| Radiation | Scattering type: x-ray |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| Reflection | Resolution: 2.9→30 Å / Num. obs: 157185 / % possible obs: 32 % / Observed criterion σ(I): 0 |
| Reflection | *PLUS Num. measured all: 412714 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Rfactor Rwork: 0.297 / Highest resolution: 2.9 Å Details: STEREOCHEMICAL CONSTRAINTS BASED ON PARAM19 AND TOP19 FILES USED IN X-PLOR VERSIONS PREVIOUS TO VERSION 3.1 ATOMIC MODELS FOR THE VIRUS AND THE DRUG WERE BUILT TO FIT ICOSAHEDRALLY ...Details: STEREOCHEMICAL CONSTRAINTS BASED ON PARAM19 AND TOP19 FILES USED IN X-PLOR VERSIONS PREVIOUS TO VERSION 3.1 ATOMIC MODELS FOR THE VIRUS AND THE DRUG WERE BUILT TO FIT ICOSAHEDRALLY CONSTRAINED 'FO' MAP USING THE GRAPHICS PROGRAM FRODO (JONES, 1978) MODIFIED TO INCORPORATE T = 1 ICOSAHEDRAL SYMMETRY. ATOMIC MODELS WERE OPTIMIZED WITH RESPECT TO THIS MAP BY A PSEUDO-REAL-SPACE REFINEMENT PROCEDURE, MINIMIZING A RESIDUAL WITH A STEREOCHEMICAL AND A CRYSTALLOGRAPHIC COMPONENT. THE GRADIENT OF THE STEREOCHEMICAL COMPONENT WAS PROVIDED BY THE X-PLOR PROGRAM (A. BRUNGER, X-PLOR VERSION 2.1 YALE UNIVERSITY 1990). THE CRYSTALLOGRAPHIC COMPONENT AND ITS GRADIENT WERE EVALUATED OVER THE VOLUME OF AN ARBITRARY PSEUDO-CELL (THE PROTOMER BOX) WHICH IS SUFFICIENTLY LARGE TO COMFORTABLY ENCLOSE A COMPLETE CHEMICALLY CONTINUOUS POLIOVIRUS PROTOMER, TOGETHER WITH WHATEVER FRAGMENTS OF SYMMETRY-RELATED PROTOMERS HAPPEN TO LIE SUFFICIENTLY CLOSE TO THE BOX TO CONTRIBUTE TO IT. THIS REFINEMENT SEEKS TO MINIMIZE THE DISCREPANCY BETWEEN THE 'PHASED' FOURIER TRANSFORMS OF MODEL-BASED ELECTRON DENSITY AND AUTHENTIC SYMMETRY-CONSTRAINED ELECTRON DENSITY, WHEN EACH TRANSFORM IS CALCULATED IN THE ARBITRARY PROTOMER BOX VOLUME, SCALED IN A RESOLUTION-DEPENDENT FASHION. SEE JRNL REFERENCE FOR MORE DETAILS. THE DISORDERED RESIDUES ABSENT FROM THE MODEL INCLUDE THE AMINO TERMINUS OF VP1 PRIOR TO THE RESIDUE LABELED GLN 24, 1 - 5 IN VP2, 236 - 238 IN VP3, AND THE RESIDUES LABELED 17 - 22 IN VP4. THE POLYPEPTIDE DESIGNATED IN THIS FILE AS RESIDUES 6 - 9 OF CHAIN 0 REPRESENTS A FEATURE IN THE ELECTRON DENSITY MAP WHICH APPEARS TO BE A BETA STRAND. ALTHOUGH THE SIDE CHAINS OF THIS STRAND CANNOT BE CORRELATED RELIABLY WITH THE SEQUENCE OF THE PROTEIN, THE FEATURE IS BELIEVED LIKELY TO CORRESPOND TO SOME PORTION OF THE AMINO TERMINAL EXTENSION OF VP1. THE POLYPEPTIDE DESIGNATED IN THIS FILE AS RESIDUES 6 - 9 OF CHAIN 0 REPRESENTS A FEATURE IN THE ELECTRON DENSITY MAP WHICH APPEARS TO BE A BETA STRAND. ALTHOUGH THE SIDE CHAINS OF THIS STRAND CANNOT BE CORRELATED RELIABLY WITH THE SEQUENCE OF THE PROTEIN, THE FEATURE IS BELIEVED LIKELY TO CORRESPOND TO SOME PORTION OF THE AMINO TERMINAL EXTENSION OF VP1. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Highest resolution: 2.9 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: 'PSEUDO-REAL SPACE PROCEDURE' / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor obs: 0.297 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller


























 PDBj
PDBj






