[English] 日本語
 Yorodumi
Yorodumi- PDB-1ua1: Structure of aminofluorene adduct paired opposite cytosine at the... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1ua1 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Structure of aminofluorene adduct paired opposite cytosine at the polymerase active site. | ||||||
 Components Components |
| ||||||
 Keywords Keywords | TRANSFERASE/DNA / DNA polymerase I / DNA replication / klenow fragment / protein-DNA complex / aminofluorene / aromatic amine / DNA lesion / translation replication / TRANSFERASE-DNA COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology information5'-3' exonuclease activity / 3'-5' exonuclease activity / DNA-templated DNA replication / double-strand break repair / DNA-directed DNA polymerase / DNA-directed DNA polymerase activity / DNA binding Similarity search - Function | ||||||
| Biological species |   Geobacillus stearothermophilus (bacteria) Geobacillus stearothermophilus (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2 Å MOLECULAR REPLACEMENT / Resolution: 2 Å | ||||||
 Authors Authors | Hsu, G.W. / Kiefer, J.R. / Becherel, O.J. / Fuchs, R.P.P. / Beese, L.S. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2004 Journal: J.Biol.Chem. / Year: 2004Title: Observing translesion synthesis of an aromatic amine DNA adduct by a high-fidelity DNA polymerase Authors: Hsu, G.W. / Kiefer, J.R. / Becherel, O.J. / Fuchs, R.P.P. / Beese, L.S. #1:  Journal: Cell(Cambridge,Mass.) / Year: 2004 Journal: Cell(Cambridge,Mass.) / Year: 2004Title: Structures of Mismatch Replication Errors Observed in a DNA Polymerase Authors: Johnson, S.J. / Beese, L.S. #2:  Journal: Proc.Natl.Acad.Sci.USA / Year: 2003 Journal: Proc.Natl.Acad.Sci.USA / Year: 2003Title: Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations Authors: Johnson, S.J. / Taylor, J.S. / Beese, L.S. #3:  Journal: Nature / Year: 1998 Journal: Nature / Year: 1998Title: Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal Authors: Kiefer, J.R. / Mao, C. / Braman, J.C. / Beese, L.S. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1ua1.cif.gz 1ua1.cif.gz | 154 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1ua1.ent.gz pdb1ua1.ent.gz | 114.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1ua1.json.gz 1ua1.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ua/1ua1 https://data.pdbj.org/pub/pdb/validation_reports/ua/1ua1 ftp://data.pdbj.org/pub/pdb/validation_reports/ua/1ua1 ftp://data.pdbj.org/pub/pdb/validation_reports/ua/1ua1 | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||
| Unit cell |
| ||||||||||
| Details | Exists as a monomer. One molecule per asymmetric unit |
- Components
Components
-DNA chain , 2 types, 2 molecules BC
| #1: DNA chain | Mass: 3454.258 Da / Num. of mol.: 1 / Source method: obtained synthetically |
|---|---|
| #2: DNA chain | Mass: 4160.720 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: see remark 400 |
-Protein , 1 types, 1 molecules A
| #3: Protein | Mass: 66114.742 Da / Num. of mol.: 1 / Fragment: analogous to the E. coli klenow fragment Source method: isolated from a genetically manipulated source Details: see remark 400 Source: (gene. exp.)   Geobacillus stearothermophilus (bacteria) Geobacillus stearothermophilus (bacteria)Plasmid: pet-30A(+) / Production host:  |
|---|
-Non-polymers , 3 types, 467 molecules 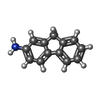




| #4: Chemical | ChemComp-AF / | ||
|---|---|---|---|
| #5: Chemical | | #6: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.75 Å3/Da / Density % sol: 54.9 % | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 290 K / Method: vapor diffusion, hanging drop / pH: 5.8 Details: Ammonium Sulfate, Magnesium Sulfate, MPD, MES, pH 5.8, VAPOR DIFFUSION, HANGING DROP, temperature 290K | ||||||||||||||||||||||||||||||||
| Components of the solutions |
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 Å ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 Å |
| Detector | Type: RIGAKU RAXIS IV / Detector: IMAGE PLATE / Date: Aug 31, 1999 / Details: dual optic mirrors |
| Radiation | Monochromator: Nickel filter / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2→35 Å / Num. all: 59431 / Num. obs: 59431 / % possible obs: 97.9 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Biso Wilson estimate: 19.8 Å2 / Rmerge(I) obs: 0.084 / Net I/σ(I): 14.9 |
| Reflection shell | Resolution: 2→2.03 Å / Rmerge(I) obs: 0.304 / Mean I/σ(I) obs: 2.4 / Num. unique all: 2485 / % possible all: 85.1 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 2→33.7 Å / Rfactor Rfree error: 0.005 / Data cutoff high absF: 2523114.46 / Data cutoff low absF: 0 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: Engh & Huber MOLECULAR REPLACEMENT / Resolution: 2→33.7 Å / Rfactor Rfree error: 0.005 / Data cutoff high absF: 2523114.46 / Data cutoff low absF: 0 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: Engh & Huber
| ||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 45.9266 Å2 / ksol: 0.366778 e/Å3 | ||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 31.9 Å2
| ||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2→33.7 Å
| ||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2→2.07 Å / Rfactor Rfree error: 0.022 / Total num. of bins used: 10
| ||||||||||||||||||||||||||||||||||||
| Xplor file |
|
 Movie
Movie Controller
Controller























 PDBj
PDBj








































