+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1jmb | ||||||
|---|---|---|---|---|---|---|---|
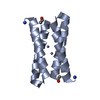
| Title | CRYSTAL STRUCTURE OF FOUR-HELIX BUNDLE MODEL | ||||||
 Components Components | PROTEIN (FOUR-HELIX BUNDLE MODEL) | ||||||
 Keywords Keywords | DE NOVO PROTEIN / ALPHA-HELICAL BUNDLE / PROTEIN DESIGN | ||||||
| Function / homology | Immunoglobulin FC, subunit C / Single alpha-helices involved in coiled-coils or other helix-helix interfaces / Up-down Bundle / Mainly Alpha / :  Function and homology information Function and homology information | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.2 Å MOLECULAR REPLACEMENT / Resolution: 2.2 Å | ||||||
 Authors Authors | Di Costanzo, L. / Geremia, S. | ||||||
 Citation Citation |  Journal: J.Am.Chem.Soc. / Year: 2001 Journal: J.Am.Chem.Soc. / Year: 2001Title: Toward the de novo design of a catalytically active helix bundle: a substrate-accessible carboxylate-bridged dinuclear metal center. Authors: Di Costanzo, L. / Wade, H. / Geremia, S. / Randaccio, L. / Pavone, V. / DeGrado, W.F. / Lombardi, A. #1:  Journal: Proc.Natl.Acad.Sci.USA / Year: 2000 Journal: Proc.Natl.Acad.Sci.USA / Year: 2000Title: Retrostructural Analysis of Metalloproteins: Application to the Design of a Minimal Model for Diiron Proteins Authors: Lombardi, A. / Summa, C.M. / Geremia, S. / Randaccio, L. / Pavone, V. / DeGrado, W.F. #2:  Journal: Curr.Opin.Struct.Biol. / Year: 1999 Journal: Curr.Opin.Struct.Biol. / Year: 1999Title: Tertiary Templates for the Design of Diiron Proteins Authors: Summa, C.M. / Lombardi, A. / Lewis, M. / DeGrado, W.F. #3:  Journal: Annu.Rev.Biochem. / Year: 1999 Journal: Annu.Rev.Biochem. / Year: 1999Title: De Novo Design and Structural Characterization of Proteins and Metalloproteins Authors: DeGrado, W.F. / Summa, C.M. / Pavone, V. / Nastri, F. / Lombardi, A. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1jmb.cif.gz 1jmb.cif.gz | 44.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1jmb.ent.gz pdb1jmb.ent.gz | 32.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1jmb.json.gz 1jmb.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1jmb_validation.pdf.gz 1jmb_validation.pdf.gz | 394 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1jmb_full_validation.pdf.gz 1jmb_full_validation.pdf.gz | 407.5 KB | Display | |
| Data in XML |  1jmb_validation.xml.gz 1jmb_validation.xml.gz | 6.4 KB | Display | |
| Data in CIF |  1jmb_validation.cif.gz 1jmb_validation.cif.gz | 8.7 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/jm/1jmb https://data.pdbj.org/pub/pdb/validation_reports/jm/1jmb ftp://data.pdbj.org/pub/pdb/validation_reports/jm/1jmb ftp://data.pdbj.org/pub/pdb/validation_reports/jm/1jmb | HTTPS FTP |
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein/peptide | Mass: 5828.813 Da / Num. of mol.: 3 / Source method: obtained synthetically / Details: THIS PROTEIN WAS CHEMICALLY SYNTHESIZED #2: Chemical | #3: Chemical | #4: Water | ChemComp-HOH / | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.38 Å3/Da / Density % sol: 48.39 % | |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 279 K / Method: vapor diffusion, hanging drop / pH: 7.5 Details: PEG 200, DMSO, Mn(CH3COO)2 , TRIS, pH 7.50, VAPOR DIFFUSION, HANGING DROP, temperature 279K | |||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 277 K / pH: 7.5 | |||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ELETTRA ELETTRA  / Beamline: 5.2R / Wavelength: 1.2 / Beamline: 5.2R / Wavelength: 1.2 |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: May 1, 2000 / Details: MIRROR |
| Radiation | Monochromator: SI(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.2 Å / Relative weight: 1 |
| Reflection | Resolution: 2.2→20 Å / Num. all: 18643 / Num. obs: 7562 / % possible obs: 87.3 % / Observed criterion σ(F): 0 / Redundancy: 2.5 % / Biso Wilson estimate: 27 Å2 / Rmerge(I) obs: 0.104 / Rsym value: 0.104 / Net I/σ(I): 5.5 |
| Reflection shell | Resolution: 2.2→2.32 Å / Redundancy: 2.4 % / Rmerge(I) obs: 0.329 / Mean I/σ(I) obs: 2.3 / Rsym value: 0.329 / % possible all: 90.5 |
| Reflection | *PLUS Lowest resolution: 21.2 Å / Num. measured all: 18643 |
| Reflection shell | *PLUS % possible obs: 90.5 % / Mean I/σ(I) obs: 2.2 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: THEORETICAL MODEL Resolution: 2.2→20 Å / Isotropic thermal model: Isotropic / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: Engh & Huber
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 41.8 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.2→20 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: REFMAC / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Num. reflection obs: 7186 / % reflection Rfree: 5 % / Rfactor obs: 0.247 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS Biso mean: 41.8 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS Type: p_angle_d / Dev ideal: 4.3 |
 Movie
Movie Controller
Controller














 PDBj
PDBj





