[English] 日本語
 Yorodumi
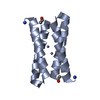
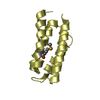
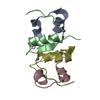
Yorodumi- PDB-1ovu: CRYSTAL STRUCTURE OF FOUR-HELIX BUNDLE MODEL di-Co(II)-DF1-L13A (... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1ovu | ||||||
|---|---|---|---|---|---|---|---|
| Title | CRYSTAL STRUCTURE OF FOUR-HELIX BUNDLE MODEL di-Co(II)-DF1-L13A (form I) | ||||||
 Components Components | four-helix bundle model di-Co(II)-DF1-L13A (form I) | ||||||
 Keywords Keywords | DE NOVO PROTEIN / ALPHA-HELICAL BUNDLE / PROTEIN DESIGN | ||||||
| Function / homology | Immunoglobulin FC, subunit C / Single alpha-helices involved in coiled-coils or other helix-helix interfaces / Up-down Bundle / Mainly Alpha / :  Function and homology information Function and homology information | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / UNCONVENTIONAL METHOD USING THE GROUP-SUBGROUP RELATION / Resolution: 3.1 Å SYNCHROTRON / UNCONVENTIONAL METHOD USING THE GROUP-SUBGROUP RELATION / Resolution: 3.1 Å | ||||||
 Authors Authors | Di Costanzo, L. / Geremia, S. | ||||||
 Citation Citation |  Journal: J.Am.Chem.Soc. / Year: 2005 Journal: J.Am.Chem.Soc. / Year: 2005Title: Response of a designed metalloprotein to changes in metal ion coordination, exogenous ligands, and active site volume determined by X-ray crystallography. Authors: Geremia, S. / Di Costanzo, L. / Randaccio, L. / Engel, D.E. / Lombardi, A. / Nastri, F. / DeGrado, W.F. #1: Journal: Acta Crystallogr.,Sect.D / Year: 2003 Title: Phasing protein structures using the group-subgroup relation. Authors: Di Costanzo, L. / Forneris, F. / Geremia, S. / Randaccio, L. #2:  Journal: ANGEW.CHEM.INT.ED.ENGL. / Year: 2003 Journal: ANGEW.CHEM.INT.ED.ENGL. / Year: 2003Title: Sliding Helix Induced Change of Coordination Geomet Model Di-Mn(II) Protein Authors: Degrado, W.F. / Di Costanzo, L. / Geremia, S. / Lombardi, A. / Pavone, V. / Randaccio, L. #3:  Journal: J.Am.Chem.Soc. / Year: 2001 Journal: J.Am.Chem.Soc. / Year: 2001Title: Toward the De Novo Design of a Catalytically Active Helix-Bundle: A Substrate Accessible Carboxylate-Br Dinuclear Metal Center Authors: Di Costanzo, L. / Wade, H. / Geremia, S. / Randaccio, L. / Pavone, V. / Degrado, W.F. / Lombardi, A. #4:  Journal: Proc.Natl.Acad.Sci.USA / Year: 2000 Journal: Proc.Natl.Acad.Sci.USA / Year: 2000Title: Retrostructural Analysis of Metalloproteins: Application to the Design of a Minimal Model for Diiron Proteins Authors: Lombardi, A. / Summa, C.M. / Geremia, S. / Randaccio, L. / Pavone, V. / Degrado, W.F. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1ovu.cif.gz 1ovu.cif.gz | 54.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1ovu.ent.gz pdb1ovu.ent.gz | 41 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1ovu.json.gz 1ovu.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ov/1ovu https://data.pdbj.org/pub/pdb/validation_reports/ov/1ovu ftp://data.pdbj.org/pub/pdb/validation_reports/ov/1ovu ftp://data.pdbj.org/pub/pdb/validation_reports/ov/1ovu | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||||||||||||||||||||
| 2 | 
| ||||||||||||||||||||||||||||||
| 3 | 
| ||||||||||||||||||||||||||||||
| Unit cell |
| ||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments: Component-ID: 1 / Ens-ID: 1 / Beg auth comp-ID: ASP / Beg label comp-ID: ASP / End auth comp-ID: GLY / End label comp-ID: GLY / Refine code: 6 / Auth seq-ID: 1 - 48 / Label seq-ID: 2 - 49
|
- Components
Components
| #1: Protein/peptide | Mass: 5828.813 Da / Num. of mol.: 4 / Source method: obtained synthetically / Details: THIS PROTEIN WAS CHEMICALLY SYNTHESIZED. #2: Chemical | ChemComp-CO / #3: Water | ChemComp-HOH / | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.57 Å3/Da / Density % sol: 53.99 % |
|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, hanging drop / pH: 7.5 Details: CO(CH3COO)2 30 mM, buffer tris 100 mM pH 7.5, peg 400 43%, VAPOR DIFFUSION, HANGING DROP, temperature 298K, pH 7.50 |
-Data collection
| Diffraction | Mean temperature: 100 K | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ELETTRA ELETTRA  / Beamline: 5.2R / Wavelength: 1.2 / Wavelength: 1.2 Å / Beamline: 5.2R / Wavelength: 1.2 / Wavelength: 1.2 Å | |||||||||
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Sep 1, 2001 / Details: MIRROR | |||||||||
| Radiation | Monochromator: SI(111) / Protocol: 1.2 / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||
| Radiation wavelength |
| |||||||||
| Reflection | Resolution: 3.1→43.2 Å / Num. obs: 4679 / % possible obs: 97.7 % / Redundancy: 3.6 % / Rmerge(I) obs: 0.236 / Rsym value: 0.236 / Net I/σ(I): 5.9 | |||||||||
| Reflection shell | Resolution: 3.1→3.27 Å / Redundancy: 3.8 % / Rmerge(I) obs: 0.515 / Mean I/σ(I) obs: 1.9 / Rsym value: 0.515 / % possible all: 97.7 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure: UNCONVENTIONAL METHOD USING THE GROUP-SUBGROUP RELATION Resolution: 3.1→20 Å / Cor.coef. Fo:Fc: 0.889 / Cor.coef. Fo:Fc free: 0.843 / SU B: 28.119 / SU ML: 0.511 / Cross valid method: THROUGHOUT / ESU R Free: 0.593
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: BABINET MODEL WITH MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 58.981 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3.1→20 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints NCS | Ens-ID: 1 / Number: 414 / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 3.1→3.179 Å / Total num. of bins used: 20 /
|
 Movie
Movie Controller
Controller















 PDBj
PDBj




