+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1j5h | ||||||
|---|---|---|---|---|---|---|---|
| Title | Solution Structure of Apo-Neocarzinostatin | ||||||
 Components Components | Apo-Neocarzinostatin | ||||||
 Keywords Keywords | ANTIBIOTIC / Beta sandwich / IgG fold | ||||||
| Function / homology |  Function and homology information Function and homology information | ||||||
| Biological species |  Streptomyces carzinostaticus (bacteria) Streptomyces carzinostaticus (bacteria) | ||||||
| Method | SOLUTION NMR / Simulated annealing with torsion angle dynamics | ||||||
 Authors Authors | Urbaniak, M.D. / Muskett, F.W. / Finucane, M.D. / Caddick, S. / Woolfson, D.N. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 2002 Journal: Biochemistry / Year: 2002Title: Solution Structure of a Novel Chromoprotein Derived from Apo-Neocarzinostatin and a Synthetic Chromophore Authors: Urbaniak, M.D. / Muskett, F.W. / Finucane, M.D. / Caddick, S. / Woolfson, D.N. | ||||||
| History |
|
- Structure visualization
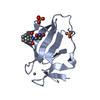
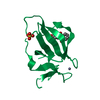
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1j5h.cif.gz 1j5h.cif.gz | 1.3 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1j5h.ent.gz pdb1j5h.ent.gz | 1 MB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1j5h.json.gz 1j5h.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/j5/1j5h https://data.pdbj.org/pub/pdb/validation_reports/j5/1j5h ftp://data.pdbj.org/pub/pdb/validation_reports/j5/1j5h ftp://data.pdbj.org/pub/pdb/validation_reports/j5/1j5h | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1j5iC C: citing same article ( |
|---|---|
| Similar structure data | |
| Other databases |
|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 12227.273 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Streptomyces carzinostaticus (bacteria) Streptomyces carzinostaticus (bacteria)Plasmid: pCANTABB5 / Production host:  |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Contents: 2.2 mM u-15N 1, 25 mM phosphate pH 5.0, 0.1 mM TSP, 0.005% sodium azide w/v Solvent system: 90% H2O/10% D2O |
|---|---|
| Sample conditions | Ionic strength: 25 mM sodium phosphate / pH: 5 / Pressure: ambient / Temperature: 308 K |
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: Simulated annealing with torsion angle dynamics / Software ordinal: 1 Details: RESTRAINTS FOR STRUCTURE CALCULATION: INTRARESIDUE NOES 194, SEQUENTIAL NOES (I, I +1) 206, MEDIUM-RANGE NOES (I, I+2 - I+4) 129, LONG-RANGE NOES (I, I> 5) 303, PSI ANGLE RESTRAINTS 56, ...Details: RESTRAINTS FOR STRUCTURE CALCULATION: INTRARESIDUE NOES 194, SEQUENTIAL NOES (I, I +1) 206, MEDIUM-RANGE NOES (I, I+2 - I+4) 129, LONG-RANGE NOES (I, I> 5) 303, PSI ANGLE RESTRAINTS 56, HYDROGEN BONDS 52, DISULPHIDE BONDS 12. | ||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with favorable non-bond energy Conformers calculated total number: 100 / Conformers submitted total number: 44 |
 Movie
Movie Controller
Controller











 PDBj
PDBj HSQC
HSQC