[English] 日本語
 Yorodumi
Yorodumi- PDB-1ik3: LIPOXYGENASE-3 (SOYBEAN) COMPLEX WITH 13(S)-HYDROPEROXY-9(Z),11(E... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1ik3 | ||||||
|---|---|---|---|---|---|---|---|
| Title | LIPOXYGENASE-3 (SOYBEAN) COMPLEX WITH 13(S)-HYDROPEROXY-9(Z),11(E)-OCTADECADIENOIC ACID | ||||||
 Components Components | LIPOXYGENASE-3 | ||||||
 Keywords Keywords | OXIDOREDUCTASE / PURPLE LIPOXYGENASE / FE(III) COMPLEX / INTERM | ||||||
| Function / homology |  Function and homology information Function and homology informationlinoleate 9S-lipoxygenase / linoleate 9S-lipoxygenase activity / oxylipin biosynthetic process / lipid oxidation / oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen / fatty acid biosynthetic process / metal ion binding / cytoplasm Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2 Å MOLECULAR REPLACEMENT / Resolution: 2 Å | ||||||
 Authors Authors | Skrzypczak-Jankun, E. / Funk Jr., M.O. | ||||||
 Citation Citation |  Journal: J.Am.Chem.Soc. / Year: 2001 Journal: J.Am.Chem.Soc. / Year: 2001Title: Three-dimensional structure of a purple lipoxygenase. Authors: Skrzypczak-Jankun, E. / Bross, R.A. / Carroll, R.T. / Dunham, W.R. / Funk, M.O. #1:  Journal: Proteins / Year: 1997 Journal: Proteins / Year: 1997Title: Structure of Soybean Lipoxygenase L3 and a Comparison with its L1 Isoenzyme Authors: Skrzypczak-Jankun, E. / Amzel, L.M. / Kroa, B.A. / Funk Jr., M.O. #2:  Journal: Biochemistry / Year: 1998 Journal: Biochemistry / Year: 1998Title: Structural and Thermochemical Characterization of Lipoxygenase-Catechol Complexes Authors: Pham, C. / Jankun, J. / Skrzypczak-Jankun, E. / Flowers II, R.A. / Funk Jr., M.O. #3:  Journal: INT.J.MOL.MED. / Year: 2000 Journal: INT.J.MOL.MED. / Year: 2000Title: Curcumin Inhibits Lipoxygenase by Binding to its Central Cavity: Theoretical and X-Ray Evidence. Authors: Skrzypczak-Jankun, E. / Mccabe, N.P. / Selman, S.H. / Jankun, J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1ik3.cif.gz 1ik3.cif.gz | 195.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1ik3.ent.gz pdb1ik3.ent.gz | 152.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1ik3.json.gz 1ik3.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1ik3_validation.pdf.gz 1ik3_validation.pdf.gz | 593.1 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1ik3_full_validation.pdf.gz 1ik3_full_validation.pdf.gz | 647.8 KB | Display | |
| Data in XML |  1ik3_validation.xml.gz 1ik3_validation.xml.gz | 49.4 KB | Display | |
| Data in CIF |  1ik3_validation.cif.gz 1ik3_validation.cif.gz | 67.2 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ik/1ik3 https://data.pdbj.org/pub/pdb/validation_reports/ik/1ik3 ftp://data.pdbj.org/pub/pdb/validation_reports/ik/1ik3 ftp://data.pdbj.org/pub/pdb/validation_reports/ik/1ik3 | HTTPS FTP |
-Related structure data
| Related structure data |  1lnhS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| Unit cell |
| |||||||||
| Components on special symmetry positions |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 96919.000 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|
-Non-polymers , 6 types, 534 molecules 
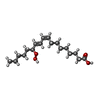









| #2: Chemical | ChemComp-FE / |
|---|---|
| #3: Chemical | ChemComp-13S / |
| #4: Chemical | ChemComp-13R / |
| #5: Chemical | ChemComp-9OH / ( |
| #6: Chemical | ChemComp-11O / ( |
| #7: Water | ChemComp-HOH / |
-Details
| Has protein modification | N |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 2 X-RAY DIFFRACTION / Number of used crystals: 2 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.5 Å3/Da / Density % sol: 50 % | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 296 K / Method: vapor diffusion, sitting drop / pH: 5.3 Details: PEG 8000, citrate-phosphate buffer, sodium azide, pH 5.3, VAPOR DIFFUSION, SITTING DROP, temperature 296K | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 23 ℃Details: drop1:drop2:drop3:drop4=3:6:1:2ratio, Skrzypczak-Jankun, E., (1997) Proteins: Struct.,Funct., Genet., 29, 15. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 296 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 |
| Detector | Type: RIGAKU RAXIS IV / Detector: IMAGE PLATE / Date: Nov 15, 1997 / Details: MIRRORS |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2→40 Å / Num. all: 48693 / Num. obs: 41035 / % possible obs: 78 % / Observed criterion σ(F): 2 / Observed criterion σ(I): 1 / Redundancy: 3 % / Biso Wilson estimate: 33 Å2 / Rmerge(I) obs: 0.064 / Net I/σ(I): 11.5 |
| Reflection shell | Resolution: 2→2.07 Å / Redundancy: 2 % / Rmerge(I) obs: 0.352 / Mean I/σ(I) obs: 1.3 / % possible all: 68 |
| Reflection | *PLUS % possible obs: 78 % / Num. measured all: 107634 |
| Reflection shell | *PLUS % possible obs: 68 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1LNH, PROTEIN ONLY Resolution: 2→40 Å / Rfactor Rfree error: 0.02 / Data cutoff high absF: 1000000 / Data cutoff low absF: 0.01 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 2 / σ(I): 1 / Stereochemistry target values: Engh & Huber Details: DISORDERED REGIONS 1-8 AND 33-45 WERE OMITTED FROM CALCULATIONS. There are four ligands in the coordinate section: 13S, 13R, 9OH, and 11O. 13S is alternate conformation A, occupancy 1.00. ...Details: DISORDERED REGIONS 1-8 AND 33-45 WERE OMITTED FROM CALCULATIONS. There are four ligands in the coordinate section: 13S, 13R, 9OH, and 11O. 13S is alternate conformation A, occupancy 1.00. 13R is alternate conformation B, occupancy 0.00. 9OH is alternate conformation C, occupancy 0.00. 11O is alternate conformation D, occupancy 0.00. The author refined both R and S conformations of the 13-hydroperoxy-9(Z),11(E)-octadecadienoic acid and the S came with lower discrepancy factor. Also in the soaking solution S was present as 86% versus only 14% of R. However it DOES NOT mean that the occupancy was 0.86 and 0.14 because lipoxygenase isozyme L3 turns 60% of R and 40% of S in its catalytic reaction, so it is difficult to judge what was the REAL occupancy for these two 13-S,R-isomers, because of the enzyme different affinity to them. Please see the primary citation for more details. The Rvalues for the S and R isomers are the following: for S-isomer R=0.196, Rfree=0.296; for R-isomer R=0.204, Rfree=0.302. The other two compounds 9OH and 11O were modelled to the density, no refinement, no energy minimization. The structure is metastable and it is highly possible for substrates and products to be there simultaneously.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 35.5 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.2 Å / Luzzati d res low obs: 10 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2→40 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 10
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Version: 3.851 / Classification: refinement X-PLOR / Version: 3.851 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 2 Å / Lowest resolution: 40 Å / σ(F): 2 / % reflection Rfree: 10 % | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS Biso mean: 35.5 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS % reflection Rfree: 4 % |
 Movie
Movie Controller
Controller















 PDBj
PDBj




