[English] 日本語
 Yorodumi
Yorodumi- PDB-1hu9: LIPOXYGENASE-3 (SOYBEAN) COMPLEX WITH 4-HYDROPEROXY-2-METHOXY-PHENOL -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1hu9 | ||||||
|---|---|---|---|---|---|---|---|
| Title | LIPOXYGENASE-3 (SOYBEAN) COMPLEX WITH 4-HYDROPEROXY-2-METHOXY-PHENOL | ||||||
 Components Components | LIPOXYGENASE-3 | ||||||
 Keywords Keywords | OXIDOREDUCTASE / METALLOPROTEIN / FE(III) COMPLEX / PURPLE LIPOXYGENASE / ENZYME INHIBITOR | ||||||
| Function / homology |  Function and homology information Function and homology informationlinoleate 9S-lipoxygenase / linoleate 9S-lipoxygenase activity / oxylipin biosynthetic process / lipid oxidation / oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen / fatty acid biosynthetic process / metal ion binding / cytoplasm Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.2 Å MOLECULAR REPLACEMENT / Resolution: 2.2 Å | ||||||
 Authors Authors | Zhou, K. / Skrzypczak-Jankun, E. / McCabe, N.P. / Selman, S.H. / Jankun, J. | ||||||
 Citation Citation |  Journal: INT.J.MOL.MED. / Year: 2003 Journal: INT.J.MOL.MED. / Year: 2003Title: Structure of curcumin in complex with lipoxygenase and its significance in cancer. Authors: Skrzypczak-Jankun, E. / Zhou, K. / McCabe, N.P. / Selman, S.H. / Jankun, J. #1:  Journal: Int.J.Mol.Med. / Year: 2000 Journal: Int.J.Mol.Med. / Year: 2000Title: Curcumin Inhibits Lipooxygenase by Binding to its Center Cavity: Theoretical and X-Ray Evidence Authors: Skrzypczak-Jankun, E. / McCabe, N.P. / Selman, S.H. / Jankun, J. #2:  Journal: Proteins / Year: 1997 Journal: Proteins / Year: 1997Title: Structure of Soybean Lipoxygenase L3 and a Comparison with its L1 Isoenzyme Authors: Skrzypczak-Jankun, E. / Amzel, L.M. / Kroa, B.A. / Funk Jr., M.O. #3:  Journal: Biochemistry / Year: 1998 Journal: Biochemistry / Year: 1998Title: Structural and Thermochemical Characterization of Lipoxygenase-Catechol Complexes Authors: Pham, C. / Jankun, J. / Skrzypczak-Jankun, E. / Flowers II, R.A. / Funk Jr., M.O. #4:  Journal: Biochemistry / Year: 1994 Journal: Biochemistry / Year: 1994Title: Position 713 is Critical for Catalysis But not Iron Binding in Soybean Lipoxygenase 3 Authors: Kramer, J.A. / Johnson, K.R. / Dunham, W.R. / Sands, R.H. / Funk Jr., M.O. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1hu9.cif.gz 1hu9.cif.gz | 190.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1hu9.ent.gz pdb1hu9.ent.gz | 149.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1hu9.json.gz 1hu9.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/hu/1hu9 https://data.pdbj.org/pub/pdb/validation_reports/hu/1hu9 ftp://data.pdbj.org/pub/pdb/validation_reports/hu/1hu9 ftp://data.pdbj.org/pub/pdb/validation_reports/hu/1hu9 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1lnhS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 96919.000 Da / Num. of mol.: 1 / Source method: isolated from a natural source Details: COMPLEX WITH 4-HYDROPEROXY-2-METHOXY-PHENOL A PHOTODEGREDATION PRODUCT OF CURCUMIN Source: (natural)  |
|---|---|
| #2: Chemical | ChemComp-FE / |
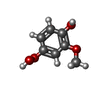
| #3: Chemical | ChemComp-4HM / |
| #4: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 2 X-RAY DIFFRACTION / Number of used crystals: 2 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.5 Å3/Da / Density % sol: 50 % |
|---|---|
| Crystal grow | Temperature: 296 K / Method: vapor diffusion, sitting drop / pH: 5.3 Details: PEG 8000, phosphate-citrate buffer, sodium azide, pH 5.3, VAPOR DIFFUSION, SITTING DROP, temperature 296K |
-Data collection
| Diffraction | Mean temperature: 295 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 |
| Detector | Type: RIGAKU RAXIS IV / Detector: IMAGE PLATE / Date: Aug 1, 1999 / Details: MIRRORS |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2.2→40 Å / Num. all: 45106 / Num. obs: 45106 / % possible obs: 95.5 % / Observed criterion σ(F): 2 / Observed criterion σ(I): 1 / Redundancy: 3 % / Biso Wilson estimate: 29.5 Å2 / Rmerge(I) obs: 0.07 / Net I/σ(I): 13.1 |
| Reflection shell | Resolution: 2.2→2.28 Å / Redundancy: 2 % / Rmerge(I) obs: 0.331 / Mean I/σ(I) obs: 1 / Num. unique all: 4304 / % possible all: 90.9 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1LNH Resolution: 2.2→40 Å / Rfactor Rfree error: 0.02 / Data cutoff high absF: 100000 / Data cutoff low absF: 0.01 / Isotropic thermal model: isotropic / Cross valid method: THROUGHOUT / σ(F): 2 / σ(I): 1 / Stereochemistry target values: Engh & Huber Details: DISORDERED REGIONS 1-8 WERE OMITTED FROM CALCULATIONS. DISORDERED REGIONS 33-45 WERE INCLUDED IN THE REFINEMENT, HOWEVER, THE HIGH B-FACTOR INDICATE DISORDER AND LOW RELIABILITY FOR THIS ...Details: DISORDERED REGIONS 1-8 WERE OMITTED FROM CALCULATIONS. DISORDERED REGIONS 33-45 WERE INCLUDED IN THE REFINEMENT, HOWEVER, THE HIGH B-FACTOR INDICATE DISORDER AND LOW RELIABILITY FOR THIS LOOP. THE IRON BINDING SITE CONSISTS OF HIS 518, HIS 523, HIS 709 ASN 713, ILE 857 (C-TERMINAL) AND 4HM 861.THESE LIGANDS ARE ALL COVALENTLY BOUND TO IRON FE 858 IN AN OCTAHEDRAL COORDINATION.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 35.2 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.24 Å / Luzzati d res low obs: 5 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.2→40 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 12
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
|
 Movie
Movie Controller
Controller

















 PDBj
PDBj