[English] 日本語
 Yorodumi
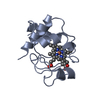
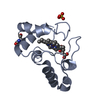
Yorodumi- PDB-1hh7: REFINED CRYSTAL STRUCTURE OF CYTOCHROME C2 FROM RHODOPSEUDOMONAS ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1hh7 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | REFINED CRYSTAL STRUCTURE OF CYTOCHROME C2 FROM RHODOPSEUDOMONAS PALUSTRIS AT 1.4 ANGSTROM RESOLUTION | |||||||||
 Components Components | CYTOCHROME C2 | |||||||||
 Keywords Keywords | ELECTRON TRANSPORT / ELECTRON CARRIER / HEME PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationphotosynthesis / electron transfer activity / heme binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  RHODOPSEUDOMONAS PALUSTRIS (phototrophic) RHODOPSEUDOMONAS PALUSTRIS (phototrophic) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.4 Å MOLECULAR REPLACEMENT / Resolution: 1.4 Å | |||||||||
 Authors Authors | Garau, G. / Geremia, S. | |||||||||
 Citation Citation |  Journal: Acta Crystallogr.,Sect.D / Year: 2000 Journal: Acta Crystallogr.,Sect.D / Year: 2000Title: Crystallization and Preliminary X-Ray Analysis of Two Ph-Dependent Forms of Cytochrome C2 from Rhodopseudomonas Palustris Authors: Garau, G. / Geremia, S. / Randaccio, L. / Vaccari, L. / Viezzoli, M.S. #1:  Journal: J.Mol.Biol. / Year: 1995 Journal: J.Mol.Biol. / Year: 1995Title: Refined Crystal Structure of Ferrocytochrome C2 from Rhodopseudomonas Viridis at 1.6 A Resolution Authors: Sogabe, S. / Miki, K. #2: Journal: Nature / Year: 1979 Title: Cytochrome C2 Sequence Variation Among the Recognised Species of Purple Nonsulphur Photosynthetic Bacteria Authors: Ambler, R.P. / Daniel, M. / Hermoso, J. / Meyer, T.E. / Bartsch, R.G. / Kamen, M.D. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1hh7.cif.gz 1hh7.cif.gz | 59.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1hh7.ent.gz pdb1hh7.ent.gz | 39.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1hh7.json.gz 1hh7.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/hh/1hh7 https://data.pdbj.org/pub/pdb/validation_reports/hh/1hh7 ftp://data.pdbj.org/pub/pdb/validation_reports/hh/1hh7 ftp://data.pdbj.org/pub/pdb/validation_reports/hh/1hh7 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2c2cS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein | Mass: 12186.983 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  RHODOPSEUDOMONAS PALUSTRIS (phototrophic) / Strain: 42 OL / References: UniProt: P00091 RHODOPSEUDOMONAS PALUSTRIS (phototrophic) / Strain: 42 OL / References: UniProt: P00091 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
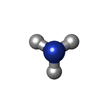
| #2: Chemical | ChemComp-HEC / | ||||||||
| #3: Chemical | | #4: Chemical | ChemComp-NH3 / | #5: Water | ChemComp-HOH / | Has protein modification | Y | Sequence details | PCA 1: GLN 1 HAS BEEN CYCLIZED TO PYRROLIDON | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.18 Å3/Da / Density % sol: 61.3 % | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 9 / Details: 60% AMMONIUM SULPHATE, 0.1 M TRIS PH 9.0 | ||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 291 K / pH: 6 / Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ELETTRA ELETTRA  / Beamline: 5.2R / Wavelength: 1 / Beamline: 5.2R / Wavelength: 1 |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Apr 23, 1999 / Details: MIRRORS |
| Radiation | Monochromator: SI(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 1.4→56 Å / Num. obs: 31246 / % possible obs: 99.8 % / Observed criterion σ(I): 0 / Redundancy: 12 % / Rmerge(I) obs: 0.096 / Net I/σ(I): 5.9 |
| Reflection shell | Resolution: 1.4→1.47 Å / Redundancy: 11.7 % / Rmerge(I) obs: 0.507 / Mean I/σ(I) obs: 5.9 / % possible all: 99.8 |
| Reflection | *PLUS Highest resolution: 1.4 Å / Lowest resolution: 56 Å / Observed criterion σ(I): 0 / Num. measured all: 394132 |
| Reflection shell | *PLUS % possible obs: 99.8 % |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 2C2C Resolution: 1.4→56 Å / Num. parameters: 4681 / Num. restraintsaints: 3727 / Cross valid method: FREE R-VALUE / σ(F): 0 Details: ANISOTROPIC REFINEMENT FOR IRON AND ALL SULFUR ATOMS CHEMICALLY EQUIVALENT BONDS AND ANGLE DISTANCES IN HEME RESTRAINED TO BE EQUAL WITHOUT TARGET VALUES. NO GEOMETRIC OR ADP RESTRAINTS APPLIED TO IRON ATOM.
| |||||||||||||||||||||||||||||||||
| Refine analyze | Num. disordered residues: 1 | |||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.4→56 Å
| |||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||
| Software | *PLUS Name: SHELX / Version: VERSION 97-1 / Classification: refinement | |||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller














 PDBj
PDBj