[English] 日本語
 Yorodumi
Yorodumi- PDB-1fj0: STRUCTURE DETERMINATION OF THE FERRICYTOCHROME C2 FROM RHODOPSEUD... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1fj0 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | STRUCTURE DETERMINATION OF THE FERRICYTOCHROME C2 FROM RHODOPSEUDOMONAS PALUSTRIS | ||||||||||||
 Components Components | CYTOCHROME C2 | ||||||||||||
 Keywords Keywords | ELECTRON TRANSPORT / ELECTRON CARRIER / HEME PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationphotosynthesis / electron transfer activity / heme binding / metal ion binding Similarity search - Function | ||||||||||||
| Biological species |  Rhodopseudomonas palustris (phototrophic) Rhodopseudomonas palustris (phototrophic) | ||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.7 Å MOLECULAR REPLACEMENT / Resolution: 1.7 Å | ||||||||||||
 Authors Authors | Geremia, S. | ||||||||||||
 Citation Citation |  Journal: Protein Sci. / Year: 2002 Journal: Protein Sci. / Year: 2002Title: Cleavage of the iron-methionine bond in c-type cytochromes: Crystal structure of oxidized and reduced cytochrome c(2) from Rhodopseudomonas palustris and its ammonia complex. Authors: Geremia, S. / Garau, G. / Vaccari, L. / Sgarra, R. / Viezzoli, M.S. / Calligaris, M. / Randaccio, L. #1:  Journal: To be Published Journal: To be PublishedTitle: Crystallization and Preliminary X-Ray Analysis of Two pH-Dependent Forms of Cytochrome C2 from Rhodopseudomonas Palustris Authors: Garau, G. / Geremia, S. / Randaccio, L. / Vaccari, L. / Viezzoli, M.S. #2:  Journal: Nature / Year: 1979 Journal: Nature / Year: 1979Title: Cytochrome C2 Sequence Variation Among the Recognised Species of Purple Nonsulphur Photosynthetic Bacteria Authors: Ambler, R.P. / Daniel, M. / Hermoso, J. / Meyer, T.E. / Bartsch, R.G. / Kamen, M.D. #3:  Journal: J.Mol.Biol. / Year: 1995 Journal: J.Mol.Biol. / Year: 1995Title: Refined Crystal Structure of Ferrocytochrome C2 From Rhodopseudomonas Viridis at 1.6 A Resolution Authors: Sogabe, S. / Miki, K. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1fj0.cif.gz 1fj0.cif.gz | 117.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1fj0.ent.gz pdb1fj0.ent.gz | 91.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1fj0.json.gz 1fj0.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/fj/1fj0 https://data.pdbj.org/pub/pdb/validation_reports/fj/1fj0 ftp://data.pdbj.org/pub/pdb/validation_reports/fj/1fj0 ftp://data.pdbj.org/pub/pdb/validation_reports/fj/1fj0 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1i8oC  1i8pC  2c2cS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 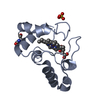
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| 4 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 12186.983 Da / Num. of mol.: 4 / Source method: isolated from a natural source / Source: (natural)  Rhodopseudomonas palustris (phototrophic) / Strain: 42 OL / References: UniProt: P00091 Rhodopseudomonas palustris (phototrophic) / Strain: 42 OL / References: UniProt: P00091#2: Chemical | #3: Chemical | ChemComp-HEC / #4: Chemical | ChemComp-GOL / | #5: Water | ChemComp-HOH / | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.46 Å3/Da / Density % sol: 44 % | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 4.4 Details: ammonium sulfate, ferricyanide, pH 4.4, VAPOR DIFFUSION, HANGING DROP, temperature 293.0K | ||||||||||||||||||||||||||||||
| Crystal grow | *PLUS pH: 6 | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ELETTRA ELETTRA  / Beamline: 5.2R / Wavelength: 1 / Beamline: 5.2R / Wavelength: 1 |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Apr 23, 1999 / Details: mirrors |
| Radiation | Monochromator: SI 111 / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 1.7→50 Å / Num. all: 194662 / Num. obs: 51959 / % possible obs: 99.9 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 3.7 % / Biso Wilson estimate: 13.42 Å2 / Rmerge(I) obs: 0.087 / Net I/σ(I): 11 |
| Reflection shell | Resolution: 1.7→1.79 Å / Redundancy: 3.7 % / Rmerge(I) obs: 0.358 / Mean I/σ(I) obs: 3.6 / Num. unique all: 7539 / % possible all: 99.7 |
| Reflection | *PLUS Num. measured all: 194662 |
| Reflection shell | *PLUS % possible obs: 99.7 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 2c2c Resolution: 1.7→50 Å / SU B: 2.19354 / SU ML: 0.07236 / σ(F): 0 / σ(I): 0 / ESU R: 0.11194 / ESU R Free: 0.11096 / Stereochemistry target values: Engh & Huber Details: The following NCS restraints were released: main-chain and side-chain of THR94, PHE95 and side-chain of LYS11, ASP20, LYS21, GLU53, LYS76, LYS96, LYS 114. The NCS restraints are as follows: ...Details: The following NCS restraints were released: main-chain and side-chain of THR94, PHE95 and side-chain of LYS11, ASP20, LYS21, GLU53, LYS76, LYS96, LYS 114. The NCS restraints are as follows: Delta 1- RMS:0.076, Sigma:0.500, Delta 2- RMS:0.101, Sigma 0.500, Delta 3- RMS:0.056, Sigma:0.500, Delta 4- RMS:0.045, Sigma:0.500.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 16.44 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.7→50 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: REFMAC / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 1.7 Å / Lowest resolution: 20 Å / σ(F): 0 / % reflection Rfree: 5 % / Rfactor obs: 0.178 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS Type: p_bond_d / Dev ideal: 0.01 |
 Movie
Movie Controller
Controller














 PDBj
PDBj














