[English] 日本語
 Yorodumi
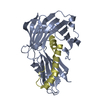
Yorodumi- PDB-1glv: THREE-DIMENSIONAL STRUCTURE OF THE GLUTATHIONE SYNTHETASE FROM ES... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1glv | ||||||
|---|---|---|---|---|---|---|---|
| Title | THREE-DIMENSIONAL STRUCTURE OF THE GLUTATHIONE SYNTHETASE FROM ESCHERICHIA COLI B AT 2.0 ANGSTROMS RESOLUTION | ||||||
 Components Components | GLUTATHIONE SYNTHASE | ||||||
 Keywords Keywords | GLUTATHIONE BIOSYNTHESIS LIGASE | ||||||
| Function / homology |  Function and homology information Function and homology informationglutathione synthase / glutathione synthase activity / glutathione biosynthetic process / protein homotetramerization / magnesium ion binding / ATP binding / identical protein binding / cytosol / cytoplasm Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / Resolution: 2.7 Å X-RAY DIFFRACTION / Resolution: 2.7 Å | ||||||
 Authors Authors | Yamaguchi, H. / Kato, H. / Tanaka, T. / Katsube, Y. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1993 Journal: J.Mol.Biol. / Year: 1993Title: Three-dimensional structure of the glutathione synthetase from Escherichia coli B at 2.0 A resolution. Authors: Yamaguchi, H. / Kato, H. / Hata, Y. / Nishioka, T. / Kimura, A. / Oda, J. / Katsube, Y. #1:  Journal: J.Mol.Biol. / Year: 1989 Journal: J.Mol.Biol. / Year: 1989Title: Crystallization and Preliminary X-Ray Studies of Glutathione Synthetase from Escherichia Coli B Authors: Kato, H. / Yamaguchi, H. / Hata, Y. / Nishioka, T. / Katsube, Y. / Oda, J. #2:  Journal: Agric.Biol.Chem. / Year: 1989 Journal: Agric.Biol.Chem. / Year: 1989Title: Overexpression of Glutathione Synthetase in Escherichia Coli Authors: Kato, H. / Kobayashi, M. / Murata, K. / Nishioka, T. / Oda, J. #3:  Journal: J.Biol.Chem. / Year: 1988 Journal: J.Biol.Chem. / Year: 1988Title: Role of Cysteine Residues in Glutathione Synthetase from Escherichia Coli B: Chemical Modification and Oligonucleotide Site-Directed Mutagenesis Authors: Kato, H. / Tanaka, T. / Nishioka, T. / Kimura, A. / Oda, J. | ||||||
| History |
| ||||||
| Remark 700 | SHEET THE SHEET STRUCTURE OF THIS MOLECULE IS BIFURCATED. IN ORDER TO REPRESENT THIS FEATURE IN THE ...SHEET THE SHEET STRUCTURE OF THIS MOLECULE IS BIFURCATED. IN ORDER TO REPRESENT THIS FEATURE IN THE SHEET RECORDS BELOW, TWO SHEETS ARE DEFINED *S1* AND *S2*. STRANDS 1 AND 2 ARE IDENTICAL. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1glv.cif.gz 1glv.cif.gz | 70.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1glv.ent.gz pdb1glv.ent.gz | 53.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1glv.json.gz 1glv.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/gl/1glv https://data.pdbj.org/pub/pdb/validation_reports/gl/1glv ftp://data.pdbj.org/pub/pdb/validation_reports/gl/1glv ftp://data.pdbj.org/pub/pdb/validation_reports/gl/1glv | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Atom site foot note | 1: CIS PROLINE - PRO 90 2: VAL 113 - ASN 114 0.829 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION 3: MET 1 EXISTS IN TWO CONFORMATIONS. | ||||||||
| Details | THE ASYMMETRIC UNIT OF THE CRYSTAL CONTAINS ONE SUBUNIT OF THE TETRAMERIC MOLECULE. SUBUNITS IN THE TETRAMERIC MOLECULE CAN BE GENERATED BY APPLYING FOLLOWING OPERATIONS TO THE FRACTIONAL CRYSTALLOGRAPHIC COORDINATES XFRAC, YFRAC, ZFRAC. 0 1 0 XFRAC 0 XFRAC2 1 0 0 X YFRAC + 0 = YFRAC2 0 0 -1 ZFRAC 0.6666666 ZFRAC2 -1 0 0 XFRAC 1 XFRAC3 0 -1 0 X YFRAC + 1 = XFRAC3 0 0 1 ZFRAC 0 XFRAC3 0 -1 0 XFRAC 1 XFRAC4 -1 0 0 X YFRAC + 1 = YFRAC4 0 0 -1 ZFRAC 0.6666666 ZFRAC4 |
- Components
Components
| #1: Protein | Mass: 34235.305 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  |
|---|---|
| #2: Water | ChemComp-HOH / |
| Compound details | THE LOOP REGION FROM ILE 226 TO ARG 241 IN THE WILD TYPE ENZYME WERE REPLACED WITH THREE GLY ...THE LOOP REGION FROM ILE 226 TO ARG 241 IN THE WILD TYPE ENZYME WERE REPLACED WITH THREE GLY RESIDUES, GLY 226, GLY 227 AND GLY 228. GLY 228 CONNECTS WITH GLY 242. |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.75 Å3/Da / Density % sol: 55.31 % | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | *PLUS Temperature: 25 ℃ / Method: microdialysis / pH: 5.5 / Details: referred to J.Mol.Biol. 209.503-504 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Radiation | Scattering type: x-ray |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| Reflection | *PLUS Highest resolution: 2.7 Å / Num. obs: 10757 / % possible obs: 98.3 % / Num. measured all: 32629 / Rmerge(I) obs: 0.033 |
- Processing
Processing
| Software | Name: PROLSQ / Classification: refinement | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 2.7→6 Å / σ(F): 2 /
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.7→6 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: PROLSQ / Classification: refinement | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 2.7 Å / Lowest resolution: 6 Å / Num. reflection obs: 9009 / σ(F): 2 / Rfactor obs: 0.177 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller











 PDBj
PDBj




