[English] 日本語
 Yorodumi
Yorodumi- PDB-1dxw: structure of hetero complex of non structural protein (NS) of hep... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1dxw | ||||||
|---|---|---|---|---|---|---|---|
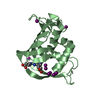
| Title | structure of hetero complex of non structural protein (NS) of hepatitis C virus (HCV) and synthetic peptidic compound | ||||||
 Components Components | SERINE PROTEASE | ||||||
 Keywords Keywords | HYDROLASE/HYDROLASE INHIBITOR / NON STRUCTURAL PROTEIN / HEPATITIS C VIRUS / SERINE PROTEASE / HYDROLASE-HYDROLASE INHIBITOR COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology informationhost cell mitochondrial membrane / host cell lipid droplet / symbiont-mediated transformation of host cell / symbiont-mediated suppression of host TRAF-mediated signal transduction / symbiont-mediated perturbation of host cell cycle G1/S transition checkpoint / host cell membrane / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT1 activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / serine-type peptidase activity / ribonucleoside triphosphate phosphatase activity ...host cell mitochondrial membrane / host cell lipid droplet / symbiont-mediated transformation of host cell / symbiont-mediated suppression of host TRAF-mediated signal transduction / symbiont-mediated perturbation of host cell cycle G1/S transition checkpoint / host cell membrane / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT1 activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / serine-type peptidase activity / ribonucleoside triphosphate phosphatase activity / host cell / channel activity / viral nucleocapsid / monoatomic ion transmembrane transport / clathrin-dependent endocytosis of virus by host cell / RNA helicase activity / host cell perinuclear region of cytoplasm / host cell endoplasmic reticulum membrane / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / ribonucleoprotein complex / symbiont-mediated activation of host autophagy / serine-type endopeptidase activity / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell nucleus / host cell plasma membrane / virion membrane / structural molecule activity / proteolysis / RNA binding / zinc ion binding / ATP binding / membrane Similarity search - Function | ||||||
| Biological species |  HEPATITIS C VIRUS HEPATITIS C VIRUS | ||||||
| Method | SOLUTION NMR / simulated annealing | ||||||
 Authors Authors | Barbato, G. / Cicero, D.O. / Cordier, F. / Narjes, F. / Gerlach, B. / Sambucini, S. / Grzesiek, S. / Matassa, V.G. / Defrancesco, R. / Bazzo, R. | ||||||
 Citation Citation |  Journal: Embo J. / Year: 2000 Journal: Embo J. / Year: 2000Title: Inhibitor Binding Induces Active Site Stabilisation of the Hcv Ns3 Protein Serine Protease Domain Authors: Barbato, G. / Cicero, D.O. / Cordier, F. / Narjes, F. / Gerlach, B. / Sambucini, S. / Grzesiek, S. / Matassa, V.G. / Defrancesco, R. / Bazzo, R. #1:  Journal: J.Mol.Biol. / Year: 1999 Journal: J.Mol.Biol. / Year: 1999Title: The Solution Structure of the N-Terminal Serine Protease Domain of the Hepatitis C Virus Ns3 Protein Provides New Insights Into its Activation and Catalytic Mechanism Authors: Barbato, G. / Cicero, D.O. / Nardi, M.C. / Steinkuhler, C. / Cortese, R. / Defrancesco, R. / Bazzo, R. #2: Journal: J.Mol.Biol. / Year: 1999 Title: Structural Characterization of the Interactions of Optimized Product Inhibitors with the N-Terminal Proteinase Domain of the Hepatitis C Virus (Hcv) Ns3 Protein by NMR and Modelling Studies Authors: Cicero, D.O. / Barbato, G. / Koch, U. / Ingallinella, P. / Bianchi, E. / Nardi, M.C. / Steinkuhler, C. / Cortese, R. / Matassa, V. / Defrancesco, R. / Pessi, A. / Bazzo, R. #3: Journal: J.Biol.Chem. / Year: 1998 Title: The Metal Binding Site of the Hepatitis C Virus Ns3 Protease Authors: Urbani, A. / Bazzo, R. / Nardi, M.C. / Cicero, D.O. / Defrancesco, R. / Steinkuhler, C. / Barbato, G. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1dxw.cif.gz 1dxw.cif.gz | 984.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1dxw.ent.gz pdb1dxw.ent.gz | 818.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1dxw.json.gz 1dxw.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/dx/1dxw https://data.pdbj.org/pub/pdb/validation_reports/dx/1dxw ftp://data.pdbj.org/pub/pdb/validation_reports/dx/1dxw ftp://data.pdbj.org/pub/pdb/validation_reports/dx/1dxw | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data | |
| Other databases |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 19634.529 Da / Num. of mol.: 1 / Fragment: SEQUENCE DATABASE RESIDUES 1027-1206 Source method: isolated from a genetically manipulated source Details: COVALENT BOND BETWEEN SER 139 OG AND ABK 190 C1 (DI-FLUORO-ABU-KETOACID) OF INHIBITOR Source: (gene. exp.)  HEPATITIS C VIRUS / Strain: BK / Plasmid: PT7-7 / Production host: HEPATITIS C VIRUS / Strain: BK / Plasmid: PT7-7 / Production host:  |
|---|---|
| #2: Chemical | ChemComp-2ZF / |
| #3: Chemical | ChemComp-ZN / |
| Has protein modification | Y |
| Sequence details | 6 RESIDUES INSERTED AT THE C TERMINAL (ALA SER LYS LYS LYS LYS) |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|---|
| NMR details | Text: THE STRUCTURE WAS DETERMINED USING TRIPLE RESONANCE EXPERIMENTS, 3D HETERONUCLEAR NOESY EXPERIMENTS, 3D AND 2D COUPLING CONSTANT MEASUREMENTS, 10% GLUCOSE C13 LABELLING FOR METHYL ...Text: THE STRUCTURE WAS DETERMINED USING TRIPLE RESONANCE EXPERIMENTS, 3D HETERONUCLEAR NOESY EXPERIMENTS, 3D AND 2D COUPLING CONSTANT MEASUREMENTS, 10% GLUCOSE C13 LABELLING FOR METHYL STEREOSPECIFIC ASSINGMENT, COMBINED USE OF SEVERAL COUPLING CONSTANTS AND ROESY SPECTROSCOPY TO DETERMINE CHI1 ANGLES. |
- Sample preparation
Sample preparation
| Details | Contents: 95% WATER/10 % D2O |
|---|---|
| Sample conditions | pH: 6.6 / Pressure: 1 atm / Temperature: 298 K |
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing / Software ordinal: 1 Details: REFINEMENT DETAILS CAN BE FOUND IN THE JRNL CITATION ABOVE. RESIDUES OF NS3 1-21 ARE NOT INCLUDED IN THE MODELS SINCE THEY ARE AFFECTED BY MOBILITY IN SOLUTION. | ||||||||||||
| NMR ensemble | Conformer selection criteria: LEAST RESTRAINT VIOLATION / Conformers calculated total number: 100 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller












 PDBj
PDBj



 X-PLOR
X-PLOR