[English] 日本語
 Yorodumi
Yorodumi- PDB-1dwk: STRUCTURE OF CYANASE WITH THE DI-ANION OXALATE BOUND AT THE ENZYM... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1dwk | ||||||
|---|---|---|---|---|---|---|---|
| Title | STRUCTURE OF CYANASE WITH THE DI-ANION OXALATE BOUND AT THE ENZYME ACTIVE SITE | ||||||
 Components Components | CYANATE HYDRATASE | ||||||
 Keywords Keywords | LYASE / CYANATE DEGRADATION / PSI / PROTEIN STRUCTURE INITIATIVE / MIDWEST CENTER FOR STRUCTURAL GENOMICS / MCSG | ||||||
| Function / homology |  Function and homology information Function and homology informationcyanate catabolic process / cyanase / cyanate hydratase activity / DNA binding Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.65 Å MOLECULAR REPLACEMENT / Resolution: 1.65 Å | ||||||
 Authors Authors | Walsh, M.A. / Otwinowski, Z. / Perrakis, A. / Anderson, P.M. / Joachimiak, A. | ||||||
 Citation Citation |  Journal: Structure / Year: 2000 Journal: Structure / Year: 2000Title: Structure of Cyanase Reveals that a Novel Dimeric and Decameric Arrangement of Subunits is Required for Formation of the Enzyme Active Site. Authors: Walsh, M.A. / Otwinowski, Z. / Perrakis, A. / Anderson, P.M. / Joachimiak, A. #1: Journal: J.Bacteriol. / Year: 1987 Title: Characterization of High-Level Expression and Sequencing of the Escherichia Coli K-12 Cyns Gene Encoding Cyanase. Authors: Sung, Y. / Anderson, P.M. / Fuchs, J.A. #2: Journal: Biochemistry / Year: 1986 Title: Kinetic Properties of Cyanase. Authors: Anderson, P.M. / Little, R.M. #3: Journal: Biochemistry / Year: 1986 Title: Interaction of Mono- and Dianions with Cyanase: Evidence for Apparent Half-Site Binding. Authors: Anderson, P.M. / Johnson, W.V. / Endrizzi, J.A. / Little, R.M. / Korte, J.J. #4: Journal: Biochemistry / Year: 1980 Title: Purification and Properties of the Inducible Enzyme Cyanase. Authors: Anderson, P.M. | ||||||
| History |
| ||||||
| Remark 700 | SHEET DETERMINATION METHOD: DSSP THE SHEET STRUCTURE OF THIS ASSEMBLY IS COMPRISED OF FIVE SHEETS ... SHEET DETERMINATION METHOD: DSSP THE SHEET STRUCTURE OF THIS ASSEMBLY IS COMPRISED OF FIVE SHEETS THAT FORM AN EQUATORIAL GIRDLE AROUND THE DECAMERIC ASSEMBLY. EACH SHEET IS MADE UP OF FOUR STRANDS FROM TWO PROTEIN CHAINS EACH CONTRIBUTING TWO STRANDS |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1dwk.cif.gz 1dwk.cif.gz | 366 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1dwk.ent.gz pdb1dwk.ent.gz | 298.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1dwk.json.gz 1dwk.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/dw/1dwk https://data.pdbj.org/pub/pdb/validation_reports/dw/1dwk ftp://data.pdbj.org/pub/pdb/validation_reports/dw/1dwk ftp://data.pdbj.org/pub/pdb/validation_reports/dw/1dwk | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1dw9SC S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||||||||||||||||||||||||||||||
| Unit cell |
| ||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS oper:
| ||||||||||||||||||||||||||||||||||||||||
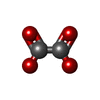
| Details | HOMODECAMER OF CYANASE CONTAINS 5 CATALYTIC SITES.IN EACH OF THESE SITES AN OXALATE ION WHICH IS ACOMPETITIVE INHIBITOR OF CYANASE IS BOUND. |
- Components
Components
| #1: Protein | Mass: 17255.377 Da / Num. of mol.: 10 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   #2: Chemical | ChemComp-SO4 / #3: Chemical | ChemComp-OXL / #4: Water | ChemComp-HOH / | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.53 Å3/Da / Density % sol: 51 % | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Method: vapor diffusion, sitting drop / pH: 7.3 Details: SELENOMETHIONINE LABELLED CRYSTALS WERE GROWN BY THE SITTING DROP METHOD OF VAPOUR DIFFUSION FROM 50% AMMONIUM SULPHATE SOLUTIONS BUFFERED WITH 50MM NAKPO4, PH = 7.3, AND IN THE PRESENCE OF ...Details: SELENOMETHIONINE LABELLED CRYSTALS WERE GROWN BY THE SITTING DROP METHOD OF VAPOUR DIFFUSION FROM 50% AMMONIUM SULPHATE SOLUTIONS BUFFERED WITH 50MM NAKPO4, PH = 7.3, AND IN THE PRESENCE OF 50 MM TRIC/HCL, PH =7.3. MICROSEEDING WITH WILD-TYPE CRYSTALS PRODUCED CRYSTALS THAT GREW TO 0.1 X 0.2 X 0.7 MM**3 OVER 5-7 DAYS. CRYSTALS WERE THEN SOAKED FOR 4 HOURS IN 5 MM SODIUM OXALATE BEFORE BEING FLASH FROZEN. | ||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 18 ℃ / Method: vapor diffusion, sitting drop | ||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 19-ID / Wavelength: 1.0335 / Beamline: 19-ID / Wavelength: 1.0335 |
| Detector | Type: ARGONNE APS-1 / Detector: CCD / Date: Nov 15, 1998 / Details: MIRROR |
| Radiation | Monochromator: SI(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.0335 Å / Relative weight: 1 |
| Reflection | Resolution: 1.65→20 Å / Num. obs: 190231 / % possible obs: 95.4 % / Redundancy: 2.3 % / Biso Wilson estimate: 13.4 Å2 / Rsym value: 0.047 / Net I/σ(I): 26.4 |
| Reflection shell | Resolution: 1.65→1.68 Å / Redundancy: 1.9 % / Mean I/σ(I) obs: 3.9 / Rsym value: 0.171 / % possible all: 79.4 |
| Reflection | *PLUS Num. measured all: 436555 / Rmerge(I) obs: 0.047 |
| Reflection shell | *PLUS % possible obs: 79.4 % / Rmerge(I) obs: 0.171 / Mean I/σ(I) obs: 4.3 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1DW9 Resolution: 1.65→20 Å / SU B: 1.07 / SU ML: 0.037 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.078 / ESU R Free: 0.088 Details: NCS RESTRAINTS NOT EMPLOYED ALTERNATIVE CONFORMATIONS WERE MODELLED FOR THE FOLLOWING AMINO ACID SIDE CHAINS CHAIN A: 25 27 31 34 60 66 78 101 132 133 CHAIN B: 10 27 66 78 101 128 CHAIN C: ...Details: NCS RESTRAINTS NOT EMPLOYED ALTERNATIVE CONFORMATIONS WERE MODELLED FOR THE FOLLOWING AMINO ACID SIDE CHAINS CHAIN A: 25 27 31 34 60 66 78 101 132 133 CHAIN B: 10 27 66 78 101 128 CHAIN C: 27 40 60 78 88 101 128 132 CHAIN D: 27 78 101 128 133 CHAIN E: 27 31 34 78 101 128 CHAIN F: 60 78 101 128 CHAIN G: 25 27 34 40 78 101 128 CHAIN H: 31 78 101 128 CHAIN I: 101 128 CHAIN J: 27 31 40 78 101 128 THIS STRUCTURE WAS DETERMINED AS PART OF THE STRUCTURAL GENOMICS INITIATIVE AT ARGONNE NATIONAL LABORATORY
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 15.8 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.65→20 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: REFMAC / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor obs: 0.139 / Rfactor Rfree: 0.178 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS Biso mean: 15.9 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller












 PDBj
PDBj