[English] 日本語
 Yorodumi
Yorodumi- PDB-1dr6: CRYSTAL STRUCTURES OF ORGANOMERCURIAL-ACTIVATED CHICKEN LIVER DIH... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1dr6 | ||||||
|---|---|---|---|---|---|---|---|
| Title | CRYSTAL STRUCTURES OF ORGANOMERCURIAL-ACTIVATED CHICKEN LIVER DIHYDROFOLATE REDUCTASE COMPLEXES | ||||||
 Components Components | DIHYDROFOLATE REDUCTASE | ||||||
 Keywords Keywords | OXIDOREDUCTASE | ||||||
| Function / homology |  Function and homology information Function and homology informationtetrahydrofolate metabolic process / response to methotrexate / dihydrofolate metabolic process / dihydrofolate reductase / dihydrofolate reductase activity / folic acid metabolic process / tetrahydrofolate biosynthetic process / one-carbon metabolic process / NADP binding / mRNA binding / mitochondrion Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / Resolution: 2.4 Å X-RAY DIFFRACTION / Resolution: 2.4 Å | ||||||
 Authors Authors | Mctigue, M.A. / Davies /II, J.F. / Kaufman, B.T. / Xuong, N.-H. / Kraut, J. | ||||||
 Citation Citation |  Journal: To be Published Journal: To be PublishedTitle: Crystal Structures of Organomercurial-Activated Chicken Liver Dihydrofolate Reductase Complexes Authors: Mctigue, M.A. / Davies II, J.F. / Kaufman, B.T. / Xuong, N.-H. / Kraut, J. #1:  Journal: Biochemistry / Year: 1990 Journal: Biochemistry / Year: 1990Title: Crystal Structures of Recombinant Human Dihydrofolate Reductase Complexed with Folate and 5-Deazafolate Authors: Davies II, J.F. / Delcamp, T.J. / Prendergast, N.J. / Ashford, V.A. / Freisheim, J.H. / Kraut, J. #2:  Journal: J.Biol.Chem. / Year: 1985 Journal: J.Biol.Chem. / Year: 1985Title: Refined Crystal Structure of Escherichia Coli and Chicken Liver Dihydrofolate Reductase Containing Bound Trimethoprim Authors: Matthews, D.A. / Bolin, J.T. / Burridge, J.M. / Filman, D.J. / Volz, K.W. / Kaufman, B.T. / Beddell, C.R. / Champness, J.N. / Stammers, D.K. / Kraut, J. #3:  Journal: J.Biol.Chem. / Year: 1985 Journal: J.Biol.Chem. / Year: 1985Title: Dihydrofolate Reductase, the Stereochemistry of Inhibitor Selectivity Authors: Matthews, D.A. / Bolin, J.T. / Burridge, J.M. / Filman, D.J. / Volz, K.W. / Kraut, J. #4:  Journal: J.Biol.Chem. / Year: 1982 Journal: J.Biol.Chem. / Year: 1982Title: Crystal Structure of Avian Dihydrofolate Reductase Containing Phenyltriazine and Nadph Authors: Volz, K.W. / Matthews, D.A. / Alden, R.A. / Freer, S.T. / Hansch, C. / Kaufman, B.T. / Kraut, J. #5:  Journal: Biochemistry / Year: 1980 Journal: Biochemistry / Year: 1980Title: Primary Structure of Chicken Liver Dihydrofolate Reductase Authors: Kumar, A.A. / Blankenship, D.T. / Kaufman, B.T. / Freisheim, J.H. | ||||||
| History |
| ||||||
| Remark 650 | HELIX RESIDUES 21-26 (NLPWPP) FORM A LEFT-HANDED POLYPROLINE HELIX. RESIDUES LYS 108 AND VAL 109 ...HELIX RESIDUES 21-26 (NLPWPP) FORM A LEFT-HANDED POLYPROLINE HELIX. RESIDUES LYS 108 AND VAL 109 PARTICIPATE IN BOTH HELIX EP AND BETA STRAND E. RESIDUES 108 AND 109 PARTICIPATE IN BOTH HELIX EP AND BETA STRAND E. | ||||||
| Remark 700 | SHEET RESIDUES ASP 110 AND MET 111 FORM A BETA-BULGE IN STRAND E. RESIDUES VAL 115 AND GLY 116 FORM ...SHEET RESIDUES ASP 110 AND MET 111 FORM A BETA-BULGE IN STRAND E. RESIDUES VAL 115 AND GLY 116 FORM A BETA-BULGE IN STRAND E. TIGHT TURN 7 (RESIDUES 162-165) DISRUPT LAST STRAND OF SHEET INTO 2 STRANDS 8 AND 9. THIS STRAND IS CONTINUOUS IN (E. COLI). A BETA BULGE IS PRESENT HERE IN (L. CASEI). RESIDUES GLU 172 AND ILE 175 PARTICIPATE IN BOTH TIGHT-TURN 8 AND BETA STRANDS 7 AND 9. |
- Structure visualization
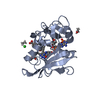
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1dr6.cif.gz 1dr6.cif.gz | 55.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1dr6.ent.gz pdb1dr6.ent.gz | 39.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1dr6.json.gz 1dr6.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/dr/1dr6 https://data.pdbj.org/pub/pdb/validation_reports/dr/1dr6 ftp://data.pdbj.org/pub/pdb/validation_reports/dr/1dr6 ftp://data.pdbj.org/pub/pdb/validation_reports/dr/1dr6 | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Atom site foot note | 1: SEE REMARK 6. 2: RESIDUES 21-26 (NLPWPP) FORM LEFT-HANDED POLYPROLINE HELIX. 3: CIS PROLINE - PRO 66 4: RESIDUES 108 AND 109 PARTICIPATE IN BOTH HELIX EP AND BETA STRAND E. 5: RESIDUES 110 AND 111 FORM A BETA-BULGE IN STRAND E. / 6: RESIDUES 115 AND 116 FORM A BETA-BULGE IN STRAND E. 7: PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION GLY 116 - GLY 117 3.713 8: TIGHT TURN 7 (RESIDUES 162-165) DISRUPT LAST STRAND OF SHEET INTO 2 STRANDS 8 AND 9. THIS STRAND IS CONTINUOUS IN E. COLI. A BETA BULGE IS PRESENT HERE IN L. CASEI. 9: RESIDUES 172 AND 175 PARTICIPATE IN BOTH TIGHT-TURN 8 AND BETA STRANDS 7 AND 9. 10: RESIDUE 200 (CALCIUM ION) LIES ON THE CRYSTALLOGRAPHIC TWO-FOLD AT ONE-HALF OCCUPANCY. | ||||||||
| Components on special symmetry positions |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 21679.932 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  |
|---|
-Non-polymers , 5 types, 78 molecules 






| #2: Chemical | ChemComp-CA / |
|---|---|
| #3: Chemical | ChemComp-NAP / |
| #4: Chemical | ChemComp-HBI / |
| #5: Chemical | ChemComp-MBO / |
| #6: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.62 Å3/Da / Density % sol: 53 % |
|---|
- Processing
Processing
| Software | Name: PROLSQ / Classification: refinement | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Rfactor obs: 0.141 / Highest resolution: 2.4 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Highest resolution: 2.4 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller




















 PDBj
PDBj







