[English] 日本語
 Yorodumi
Yorodumi- PDB-1dh3: CRYSTAL STRUCTURE OF A CREB BZIP-CRE COMPLEX REVEALS THE BASIS FO... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1dh3 | ||||||
|---|---|---|---|---|---|---|---|
| Title | CRYSTAL STRUCTURE OF A CREB BZIP-CRE COMPLEX REVEALS THE BASIS FOR CREB FAIMLY SELECTIVE DIMERIZATION AND DNA BINDING | ||||||
 Components Components |
| ||||||
 Keywords Keywords | TRANSCRIPTION/DNA / PROTEIN-DNA COMPLEX / TRANSCRIPTION-DNA COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology informationAKT phosphorylates targets in the nucleus / NGF-stimulated transcription / chemotaxis to arachidonate / response to erythropoietin / ATF4-CREB1 transcription factor complex / CREB phosphorylation / CREB1 phosphorylation through NMDA receptor-mediated activation of RAS signaling / Gastrin-CREB signalling pathway via PKC and MAPK / Estrogen-dependent nuclear events downstream of ESR-membrane signaling / positive regulation of transforming growth factor beta3 production ...AKT phosphorylates targets in the nucleus / NGF-stimulated transcription / chemotaxis to arachidonate / response to erythropoietin / ATF4-CREB1 transcription factor complex / CREB phosphorylation / CREB1 phosphorylation through NMDA receptor-mediated activation of RAS signaling / Gastrin-CREB signalling pathway via PKC and MAPK / Estrogen-dependent nuclear events downstream of ESR-membrane signaling / positive regulation of transforming growth factor beta3 production / lung saccule development / response to dehydroepiandrosterone / NCAM signaling for neurite out-growth / cAMP response element binding / response to hypobaric hypoxia / secretory granule organization / positive regulation of cardiac muscle tissue development / regulation of glial cell proliferation / lung epithelium development / inhibition of non-skeletal tissue mineralization / regulation of fibroblast proliferation / pituitary gland development / regulation of testosterone biosynthetic process / positive regulation of membrane depolarization / response to purine-containing compound / negative regulation of transcription by competitive promoter binding / hormone secretion / positive regulation of hormone secretion / mammary gland development / positive regulation of multicellular organism growth / positive regulation of osteoclast differentiation / arrestin family protein binding / cellular response to fatty acid / response to L-glutamate / response to glucagon / cellular response to hepatocyte growth factor stimulus / cellular response to insulin-like growth factor stimulus / response to morphine / histone acetyltransferase binding / regulation of cell size / type I pneumocyte differentiation / cellular response to zinc ion / positive regulation of RNA polymerase II transcription preinitiation complex assembly / cAMP/PKA signal transduction / positive regulation of fat cell differentiation / positive regulation of lipid biosynthetic process / cellular response to platelet-derived growth factor stimulus / cellular response to retinoic acid / lactation / Hsp70 protein binding / transforming growth factor beta receptor signaling pathway / cellular response to forskolin / axonogenesis / osteoclast differentiation / positive regulation of long-term synaptic potentiation / response to activity / cellular response to leukemia inhibitory factor / response to nicotine / RNA polymerase II transcription regulatory region sequence-specific DNA binding / response to cocaine / euchromatin / circadian rhythm / cellular response to nerve growth factor stimulus / visual learning / mRNA transcription by RNA polymerase II / cellular response to growth factor stimulus / RNA polymerase II transcription regulator complex / transcription coactivator binding / memory / DNA-binding transcription activator activity, RNA polymerase II-specific / transcription regulator complex / sequence-specific DNA binding / response to ethanol / RNA polymerase II-specific DNA-binding transcription factor binding / protein stabilization / ciliary basal body / positive regulation of apoptotic process / mitochondrial matrix / DNA-binding transcription factor activity / response to xenobiotic stimulus / negative regulation of gene expression / axon / centrosome / regulation of transcription by RNA polymerase II / regulation of DNA-templated transcription / negative regulation of apoptotic process / positive regulation of DNA-templated transcription / chromatin / positive regulation of transcription by RNA polymerase II / DNA binding / nucleoplasm / identical protein binding / nucleus / cytosol Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 3 Å SYNCHROTRON / Resolution: 3 Å | ||||||
 Authors Authors | Schumacher, M.A. / Goodman, R.H. / Brennan, R.G. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2000 Journal: J.Biol.Chem. / Year: 2000Title: The structure of a CREB bZIP.somatostatin CRE complex reveals the basis for selective dimerization and divalent cation-enhanced DNA binding Authors: Schumacher, M.A. / Goodman, R.H. / Brennan, R.G. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1dh3.cif.gz 1dh3.cif.gz | 60.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1dh3.ent.gz pdb1dh3.ent.gz | 41.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1dh3.json.gz 1dh3.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/dh/1dh3 https://data.pdbj.org/pub/pdb/validation_reports/dh/1dh3 ftp://data.pdbj.org/pub/pdb/validation_reports/dh/1dh3 ftp://data.pdbj.org/pub/pdb/validation_reports/dh/1dh3 | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 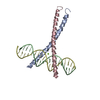
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||
| Unit cell |
|
- Components
Components
| #1: DNA chain | Mass: 6424.148 Da / Num. of mol.: 2 / Source method: obtained synthetically #2: Protein | Mass: 6686.831 Da / Num. of mol.: 2 / Fragment: RESIDUES 201-255 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   #3: Chemical | ChemComp-MG / | #4: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.42 Å3/Da / Density % sol: 64.01 % | |||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, hanging drop / pH: 5.6 Details: PEG 8000, MGCL2, (NH4)2SO4, MES, pH 5.6, VAPOR DIFFUSION, HANGING DROP, temperature 298.K | |||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions |
| |||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion | |||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 298 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRL SSRL  / Beamline: BL7-1 / Wavelength: 1.08 / Beamline: BL7-1 / Wavelength: 1.08 |
| Detector | Detector: IMAGE PLATE |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.08 Å / Relative weight: 1 |
| Reflection | Resolution: 3→10 Å / Num. all: 7436 / Num. obs: 7316 / % possible obs: 90 % / Observed criterion σ(I): 1 / Redundancy: 5.7 % / Biso Wilson estimate: 40 Å2 / Rmerge(I) obs: 0.079 / Net I/σ(I): 17 |
| Reflection shell | Resolution: 3→3.2 Å / Redundancy: 5 % / Rmerge(I) obs: 0.258 / % possible all: 95 |
| Reflection | *PLUS % possible obs: 95 % / Redundancy: 4.3 % / Rmerge(I) obs: 0.065 |
| Reflection shell | *PLUS % possible obs: 87.4 % / Mean I/σ(I) obs: 3.6 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 3→10 Å / Cross valid method: THROUGHOUT / σ(F): 2 / σ(I): 1 Details: THE FIRST AND LAST TWO RESIDUES ARE DISORDERED AND NOT MODELED. THE DENSITY IS WEAK FOR RESIDUES 334-339 IN BOTH PROTEIN CHAINS. PROCHECK REVEALED NO BAD CONTACTS AND NO RAMACHANDRAN OUTLIERS
| ||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3→10 Å
| ||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||
| Software | *PLUS Name: TNT / Classification: refinement | ||||||||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 3 Å / Lowest resolution: 10 Å / σ(F): 2 / % reflection Rfree: 10 % / Rfactor Rfree: 0.28 | ||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller






 PDBj
PDBj










































