+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9507 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of the RP region of human 26S proteasome at 4.3A | |||||||||
 Map data Map data | map of the RP region | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.3 Å | |||||||||
 Authors Authors | Huang XL / Luan B / Wu JP / Shi YG | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2016 Journal: Nat Struct Mol Biol / Year: 2016Title: An atomic structure of the human 26S proteasome. Authors: Xiuliang Huang / Bai Luan / Jianping Wu / Yigong Shi /  Abstract: We report the cryo-EM structure of the human 26S proteasome at an average resolution of 3.5 Å, allowing atomic modeling of 28 subunits in the core particle (CP) and 18 subunits in the regulatory ...We report the cryo-EM structure of the human 26S proteasome at an average resolution of 3.5 Å, allowing atomic modeling of 28 subunits in the core particle (CP) and 18 subunits in the regulatory particle (RP). The C-terminal residues of Rpt3 and Rpt5 subunits in the RP can be seen inserted into surface pockets formed between adjacent α subunits in the CP. Each of the six Rpt subunits contains a bound nucleotide, and the central gate of the CP α-ring is closed despite RP association. The six pore 1 loops in the Rpt ring are arranged similarly to a spiral staircase along the axial channel of substrate transport, which is constricted by the pore 2 loops. We also determined the cryo-EM structure of the human proteasome bound to the deubiquitinating enzyme USP14 at 4.35-Å resolution. Together, our structures provide a framework for mechanistic understanding of eukaryotic proteasome function. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9507.map.gz emd_9507.map.gz | 78.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9507-v30.xml emd-9507-v30.xml emd-9507.xml emd-9507.xml | 11.7 KB 11.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9507.png emd_9507.png | 37.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9507 http://ftp.pdbj.org/pub/emdb/structures/EMD-9507 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9507 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9507 | HTTPS FTP |
-Related structure data
| Related structure data |  9508C  9509C  9510C  9511C  9512C  5gjqC  5gjrC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9507.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9507.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map of the RP region | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
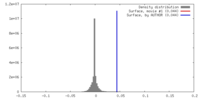
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : human 26S proteasome
| Entire | Name: human 26S proteasome |
|---|---|
| Components |
|
-Supramolecule #1: human 26S proteasome
| Supramolecule | Name: human 26S proteasome / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 2.5 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Material: COPPER / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 3.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 2s before plunging. | ||||||||||||
| Details | This sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 70.0 K |
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Number real images: 4881 / Average exposure time: 1.6 sec. / Average electron dose: 37.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)