+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-9388 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | structure of a complex | |||||||||
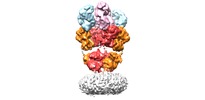
 マップデータ マップデータ | Structure of a complex | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | AMPA receptor / ligand gated ion channel / neurotransmitter / synapse / MEMBRANE PROTEIN / MEMBRANE PROTEIN-Immune System complex | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報Presynaptic depolarization and calcium channel opening / eye blink reflex / positive regulation of protein localization to basolateral plasma membrane / cerebellar mossy fiber / postsynaptic neurotransmitter receptor diffusion trapping / LGI-ADAM interactions / Trafficking of AMPA receptors / channel regulator activity / regulation of AMPA receptor activity / membrane hyperpolarization ...Presynaptic depolarization and calcium channel opening / eye blink reflex / positive regulation of protein localization to basolateral plasma membrane / cerebellar mossy fiber / postsynaptic neurotransmitter receptor diffusion trapping / LGI-ADAM interactions / Trafficking of AMPA receptors / channel regulator activity / regulation of AMPA receptor activity / membrane hyperpolarization / Synaptic adhesion-like molecules / nervous system process / protein targeting to membrane / voltage-gated calcium channel complex / neurotransmitter receptor localization to postsynaptic specialization membrane / protein heterotetramerization / spine synapse / neuromuscular junction development / dendritic spine neck / dendritic spine head / Activation of AMPA receptors / parallel fiber to Purkinje cell synapse / perisynaptic space / AMPA glutamate receptor activity / transmission of nerve impulse / ligand-gated monoatomic cation channel activity / Trafficking of GluR2-containing AMPA receptors / response to lithium ion / immunoglobulin binding / AMPA glutamate receptor complex / kainate selective glutamate receptor activity / ionotropic glutamate receptor complex / cellular response to glycine / extracellularly glutamate-gated ion channel activity / membrane depolarization / asymmetric synapse / regulation of receptor recycling / Unblocking of NMDA receptors, glutamate binding and activation / positive regulation of synaptic transmission / glutamate receptor binding / regulation of postsynaptic membrane neurotransmitter receptor levels / synaptic cleft / voltage-gated calcium channel activity / glutamate-gated receptor activity / regulation of synaptic transmission, glutamatergic / response to fungicide / cytoskeletal protein binding / ionotropic glutamate receptor binding / presynaptic active zone membrane / extracellular ligand-gated monoatomic ion channel activity / somatodendritic compartment / glutamate-gated calcium ion channel activity / cellular response to brain-derived neurotrophic factor stimulus / calcium channel regulator activity / ligand-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / dendrite membrane / dendrite cytoplasm / ionotropic glutamate receptor signaling pathway / positive regulation of synaptic transmission, glutamatergic / SNARE binding / hippocampal mossy fiber to CA3 synapse / dendritic shaft / regulation of membrane potential / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / synaptic transmission, glutamatergic / PDZ domain binding / protein tetramerization / establishment of protein localization / synaptic membrane / modulation of chemical synaptic transmission / postsynaptic density membrane / terminal bouton / Schaffer collateral - CA1 synapse / cerebral cortex development / receptor internalization / long-term synaptic potentiation / response to calcium ion / synaptic vesicle membrane / synaptic vesicle / presynapse / signaling receptor activity / amyloid-beta binding / presynaptic membrane / growth cone / scaffold protein binding / chemical synaptic transmission / protein homotetramerization / dendritic spine / perikaryon / postsynaptic membrane / neuron projection / postsynaptic density / axon / neuronal cell body / synapse / dendrite / protein-containing complex binding / protein kinase binding / glutamatergic synapse / cell surface 類似検索 - 分子機能 | |||||||||
| 生物種 |   | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 6.5 Å | |||||||||
 データ登録者 データ登録者 | Gouaux E / Zhao Y | |||||||||
 引用 引用 |  ジャーナル: Science / 年: 2019 ジャーナル: Science / 年: 2019タイトル: Architecture and subunit arrangement of native AMPA receptors elucidated by cryo-EM. 著者: Yan Zhao / Shanshuang Chen / Adam C Swensen / Wei-Jun Qian / Eric Gouaux /  要旨: Glutamate-gated AMPA receptors mediate the fast component of excitatory signal transduction at chemical synapses throughout all regions of the mammalian brain. AMPA receptors are tetrameric ...Glutamate-gated AMPA receptors mediate the fast component of excitatory signal transduction at chemical synapses throughout all regions of the mammalian brain. AMPA receptors are tetrameric assemblies composed of four subunits, GluA1-GluA4. Despite decades of study, the subunit composition, subunit arrangement, and molecular structure of native AMPA receptors remain unknown. Here we elucidate the structures of 10 distinct native AMPA receptor complexes by single-particle cryo-electron microscopy (cryo-EM). We find that receptor subunits are arranged nonstochastically, with the GluA2 subunit preferentially occupying the B and D positions of the tetramer and with triheteromeric assemblies comprising a major population of native AMPA receptors. Cryo-EM maps define the structure for S2-M4 linkers between the ligand-binding and transmembrane domains, suggesting how neurotransmitter binding is coupled to ion channel gating. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_9388.map.gz emd_9388.map.gz | 7.5 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-9388-v30.xml emd-9388-v30.xml emd-9388.xml emd-9388.xml | 30.8 KB 30.8 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_9388.png emd_9388.png | 126.5 KB | ||
| Filedesc metadata |  emd-9388.cif.gz emd-9388.cif.gz | 7.9 KB | ||
| その他 |  emd_9388_additional_1.map.gz emd_9388_additional_1.map.gz emd_9388_additional_2.map.gz emd_9388_additional_2.map.gz emd_9388_additional_3.map.gz emd_9388_additional_3.map.gz | 8.4 MB 8 MB 7.4 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9388 http://ftp.pdbj.org/pub/emdb/structures/EMD-9388 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9388 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9388 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_9388_validation.pdf.gz emd_9388_validation.pdf.gz | 338.7 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_9388_full_validation.pdf.gz emd_9388_full_validation.pdf.gz | 338.3 KB | 表示 | |
| XML形式データ |  emd_9388_validation.xml.gz emd_9388_validation.xml.gz | 6.5 KB | 表示 | |
| CIF形式データ |  emd_9388_validation.cif.gz emd_9388_validation.cif.gz | 7.6 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9388 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9388 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9388 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9388 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  6njmMC  0426C  0427C  0428C  0429C  0430C  0431C  0432C  9387C  9389C  6njlC  6njnC M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_9388.map.gz / 形式: CCP4 / 大きさ: 125 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_9388.map.gz / 形式: CCP4 / 大きさ: 125 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Structure of a complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.72 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
-追加マップ: Structure of a complex
| ファイル | emd_9388_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Structure of a complex | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-追加マップ: Structure of a complex
| ファイル | emd_9388_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Structure of a complex | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-追加マップ: Structure of a complex
| ファイル | emd_9388_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Structure of a complex | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
+全体 : Native A3A2A3A2 complex bound with MPQX
+超分子 #1: Native A3A2A3A2 complex bound with MPQX
+超分子 #2: LBD-TMD layers of native A3A2A3A2 nAMPAR complex
+超分子 #3: ATD-LBD layers of native A3A2A3A2 nAMPAR complex
+分子 #1: Glutamate receptor 3
+分子 #2: Glutamate receptor 2
+分子 #3: A'-C' auxiliary proteins
+分子 #4: Voltage-dependent calcium channel gamma-2 subunit
+分子 #5: 5B2 Fab Light Chain
+分子 #6: 5B2 Fab Heavy Chain
+分子 #7: 15F1 Fab light chain
+分子 #8: 15F1 Fab heavy chain
+分子 #11: {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquino...
+分子 #12: 2-acetamido-2-deoxy-beta-D-glucopyranose
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 4 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 緩衝液 | pH: 8 構成要素:
| ||||||||||||
| グリッド | 詳細: unspecified | ||||||||||||
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / チャンバー内温度: 295 K |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) 検出モード: SUPER-RESOLUTION / 平均電子線量: 54.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
+ 画像解析
画像解析
-原子モデル構築 1
| 精密化 | 空間: REAL / プロトコル: RIGID BODY FIT |
|---|---|
| 得られたモデル |  PDB-6njm: |
 ムービー
ムービー コントローラー
コントローラー















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)