+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9387 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | structure of a complex | |||||||||
 Map data Map data | Structure of a complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | AMPA receptor / ligand gated ion channel / neurotransmitter / synapse / MEMBRANE PROTEIN / MEMBRANE PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationCargo concentration in the ER / Presynaptic depolarization and calcium channel opening / axonal spine / positive regulation of locomotion involved in locomotory behavior / positive regulation of membrane potential / COPII-mediated vesicle transport / eye blink reflex / positive regulation of protein localization to basolateral plasma membrane / cellular response to ammonium ion / response to sucrose ...Cargo concentration in the ER / Presynaptic depolarization and calcium channel opening / axonal spine / positive regulation of locomotion involved in locomotory behavior / positive regulation of membrane potential / COPII-mediated vesicle transport / eye blink reflex / positive regulation of protein localization to basolateral plasma membrane / cellular response to ammonium ion / response to sucrose / neuron spine / myosin V binding / cerebellar mossy fiber / postsynaptic neurotransmitter receptor diffusion trapping / proximal dendrite / channel regulator activity / LGI-ADAM interactions / regulation of monoatomic ion transmembrane transport / Trafficking of AMPA receptors / regulation of AMPA receptor activity / response to arsenic-containing substance / cellular response to L-glutamate / cellular response to dsRNA / membrane hyperpolarization / dendritic spine membrane / beta-2 adrenergic receptor binding / nervous system process / long-term synaptic depression / Synaptic adhesion-like molecules / protein targeting to membrane / cellular response to peptide hormone stimulus / voltage-gated calcium channel complex / spine synapse / dendritic spine cytoplasm / dendritic spine neck / neuronal cell body membrane / cellular response to amine stimulus / response to psychosocial stress / dendritic spine head / peptide hormone receptor binding / response to morphine / neurotransmitter receptor localization to postsynaptic specialization membrane / spinal cord development / Activation of AMPA receptors / ligand-gated monoatomic cation channel activity / perisynaptic space / neuromuscular junction development / protein kinase A binding / AMPA glutamate receptor activity / Trafficking of GluR2-containing AMPA receptors / response to lithium ion / behavioral response to pain / transmission of nerve impulse / AMPA glutamate receptor clustering / cellular response to glycine / kainate selective glutamate receptor activity / adenylate cyclase binding / immunoglobulin binding / asymmetric synapse / AMPA glutamate receptor complex / response to electrical stimulus / regulation of receptor recycling / extracellularly glutamate-gated ion channel activity / ionotropic glutamate receptor complex / membrane depolarization / conditioned place preference / G-protein alpha-subunit binding / Unblocking of NMDA receptors, glutamate binding and activation / glutamate receptor binding / long-term memory / positive regulation of synaptic transmission / regulation of synaptic transmission, glutamatergic / regulation of postsynaptic membrane neurotransmitter receptor levels / positive regulation of synaptic transmission, glutamatergic / postsynaptic density, intracellular component / response to fungicide / neuronal action potential / voltage-gated calcium channel activity / cytoskeletal protein binding / extracellular ligand-gated monoatomic ion channel activity / glutamate-gated receptor activity / cellular response to brain-derived neurotrophic factor stimulus / regulation of long-term synaptic depression / synapse assembly / somatodendritic compartment / glutamate-gated calcium ion channel activity / presynaptic active zone membrane / dendrite membrane / ionotropic glutamate receptor binding / excitatory synapse / ionotropic glutamate receptor signaling pathway / dendrite cytoplasm / ligand-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / synaptic membrane / positive regulation of excitatory postsynaptic potential / dendritic shaft / hippocampal mossy fiber to CA3 synapse / SNARE binding / response to cocaine / PDZ domain binding Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.7 Å | |||||||||
 Authors Authors | Gouaux E / Zhao Y | |||||||||
 Citation Citation |  Journal: Science / Year: 2019 Journal: Science / Year: 2019Title: Architecture and subunit arrangement of native AMPA receptors elucidated by cryo-EM. Authors: Yan Zhao / Shanshuang Chen / Adam C Swensen / Wei-Jun Qian / Eric Gouaux /  Abstract: Glutamate-gated AMPA receptors mediate the fast component of excitatory signal transduction at chemical synapses throughout all regions of the mammalian brain. AMPA receptors are tetrameric ...Glutamate-gated AMPA receptors mediate the fast component of excitatory signal transduction at chemical synapses throughout all regions of the mammalian brain. AMPA receptors are tetrameric assemblies composed of four subunits, GluA1-GluA4. Despite decades of study, the subunit composition, subunit arrangement, and molecular structure of native AMPA receptors remain unknown. Here we elucidate the structures of 10 distinct native AMPA receptor complexes by single-particle cryo-electron microscopy (cryo-EM). We find that receptor subunits are arranged nonstochastically, with the GluA2 subunit preferentially occupying the B and D positions of the tetramer and with triheteromeric assemblies comprising a major population of native AMPA receptors. Cryo-EM maps define the structure for S2-M4 linkers between the ligand-binding and transmembrane domains, suggesting how neurotransmitter binding is coupled to ion channel gating. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9387.map.gz emd_9387.map.gz | 10.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9387-v30.xml emd-9387-v30.xml emd-9387.xml emd-9387.xml | 25.8 KB 25.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9387.png emd_9387.png | 91.5 KB | ||
| Filedesc metadata |  emd-9387.cif.gz emd-9387.cif.gz | 7.9 KB | ||
| Others |  emd_9387_additional_1.map.gz emd_9387_additional_1.map.gz emd_9387_additional_2.map.gz emd_9387_additional_2.map.gz emd_9387_additional_3.map.gz emd_9387_additional_3.map.gz | 7.8 MB 8.9 MB 8.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9387 http://ftp.pdbj.org/pub/emdb/structures/EMD-9387 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9387 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9387 | HTTPS FTP |
-Related structure data
| Related structure data |  6njlMC  0426C  0427C  0428C  0429C  0430C  0431C  0432C  9388C  9389C  6njmC  6njnC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9387.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9387.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of a complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.72 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
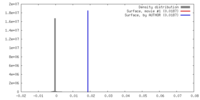
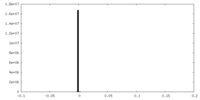
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Structure of a complex
| File | emd_9387_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of a complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
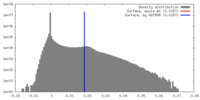
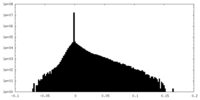
| Density Histograms |
-Additional map: Structure of a complex
| File | emd_9387_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of a complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Structure of a complex
| File | emd_9387_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of a complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Native A1A2A1A2 complex bound with MPQX
+Supramolecule #1: Native A1A2A1A2 complex bound with MPQX
+Macromolecule #1: Glutamate receptor 1
+Macromolecule #2: Glutamate receptor 2
+Macromolecule #3: A'/C' auxiliary proteins
+Macromolecule #4: Voltage-dependent calcium channel gamma-2 subunit
+Macromolecule #5: 11B8 scFv
+Macromolecule #6: 15F1 Fab light chain
+Macromolecule #7: 15F1 Fab heavy chain
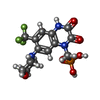
+Macromolecule #9: {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquino...
+Macromolecule #10: 2-acetamido-2-deoxy-beta-D-glucopyranose
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Details: unspecified | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 54.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-6njl: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)