+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9360 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of zebrafish Otop1 in nanodiscs | |||||||||
 Map data Map data | Structure of zfOtop1 in lipidic nanodiscs | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Proton channel / Otopetrin / MEMBRANE PROTEIN | |||||||||
| Biological species |  | |||||||||
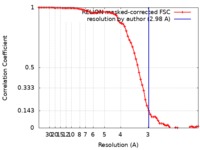
| Method | single particle reconstruction / cryo EM / Resolution: 2.98 Å | |||||||||
 Authors Authors | Saotome K / Lee WH | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2019 Journal: Nat Struct Mol Biol / Year: 2019Title: Structures of the otopetrin proton channels Otop1 and Otop3. Authors: Kei Saotome / Bochuan Teng / Che Chun Alex Tsui / Wen-Hsin Lee / Yu-Hsiang Tu / Joshua P Kaplan / Mark S P Sansom / Emily R Liman / Andrew B Ward /   Abstract: Otopetrins (Otop1-Otop3) comprise one of two known eukaryotic proton-selective channel families. Otop1 is required for otoconia formation and a candidate mammalian sour taste receptor. Here we report ...Otopetrins (Otop1-Otop3) comprise one of two known eukaryotic proton-selective channel families. Otop1 is required for otoconia formation and a candidate mammalian sour taste receptor. Here we report cryo-EM structures of zebrafish Otop1 and chicken Otop3 in lipid nanodiscs. The structures reveal a dimeric architecture, with each subunit forming 12 transmembrane helices divided into structurally similar amino (N) and carboxy (C) domains. Cholesterol-like molecules occupy various sites in Otop1 and Otop3 and occlude a central tunnel. In molecular dynamics simulations, hydrophilic vestibules formed by the N and C domains and in the intrasubunit interface between N and C domains form conduits for water entry into the membrane core, suggesting three potential proton conduction pathways. By mutagenesis, we tested the roles of charged residues in each putative permeation pathway. Our results provide a structural basis for understanding selective proton permeation and gating of this conserved family of proton channels. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9360.map.gz emd_9360.map.gz | 28.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9360-v30.xml emd-9360-v30.xml emd-9360.xml emd-9360.xml | 11.6 KB 11.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9360_fsc.xml emd_9360_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_9360.png emd_9360.png | 197.6 KB | ||
| Filedesc metadata |  emd-9360.cif.gz emd-9360.cif.gz | 5.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9360 http://ftp.pdbj.org/pub/emdb/structures/EMD-9360 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9360 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9360 | HTTPS FTP |
-Related structure data
| Related structure data |  6nf4MC  9361C  6nf6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9360.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9360.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of zfOtop1 in lipidic nanodiscs | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Zebrafish Otopetrin1 in lipidic nanodiscs
| Entire | Name: Zebrafish Otopetrin1 in lipidic nanodiscs |
|---|---|
| Components |
|
-Supramolecule #1: Zebrafish Otopetrin1 in lipidic nanodiscs
| Supramolecule | Name: Zebrafish Otopetrin1 in lipidic nanodiscs / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 65853 kDa/nm |
-Macromolecule #1: Otopetrin1
| Macromolecule | Name: Otopetrin1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 65.817 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GPVEHGGTDS MWLNKYNPAA ASSASSSSSS DAENKLFSRL KVSLTKKYPQ KNAELLSAQY GTNLLLLGVS VMLALAAQSG PVKEEHLLS FITVLMLVQL VWMLCYMIRR ERERSPVPER DAHAGASWIR GGLTMLALLS LIMDAFRIGY FVGYHSCISA A LGVYPIVH ...String: GPVEHGGTDS MWLNKYNPAA ASSASSSSSS DAENKLFSRL KVSLTKKYPQ KNAELLSAQY GTNLLLLGVS VMLALAAQSG PVKEEHLLS FITVLMLVQL VWMLCYMIRR ERERSPVPER DAHAGASWIR GGLTMLALLS LIMDAFRIGY FVGYHSCISA A LGVYPIVH ALHTISQVHF LWFHIKDVIK KYETFERFGV IHAVFTNLLL WCNGVMSETE HFMHNHRRRL IEMGYANLST VD VQPHCNC TTSVCSMFST SLYYLYPFNI EYHIFVSAML FVMWKNIGRT LDRHSNRKRR STGSTGLLLG PLGGLVALAS SVS VLVVYL IHLEKTEEMH EAAVSMFYYY GVAMMACMCV GSGTGLLVYR MENRPMDTGS NPARTLDTEL LLASSLGSWL MSWC SVVAS VAEAGQKSPS FSWTSLTYSL LLVLEKCIQN LFIVESLYRR HSEEEEDAAA PQVFSVAVPP YDGILNHGYE AHDKH REAE PAAGSHALSR KQPDAPLPAG QRLDVTPGRK RQILKNICMF LFMCNISLWI LPAFGCRPQY DNPLENETFG TSVWTT VLN VAIPLNLFYR MHSVASLFEV FRKV |
-Macromolecule #2: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 2 / Number of copies: 6 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Macromolecule #3: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 3 / Number of copies: 2 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.1 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil, UltrAuFoil, R1.2/1.3 / Material: GOLD |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)