[English] 日本語
 Yorodumi
Yorodumi- EMDB-9138: HIV-1 vaccine elicited Fab BDA1 in complex with the HIV Env trime... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9138 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | HIV-1 vaccine elicited Fab BDA1 in complex with the HIV Env trimer BG505 SOSIPv5.2 | |||||||||
 Map data Map data | HIV-1 vaccine elicited Fab BDA1 in complex with the HIV Env trimer BG505 SOSIPv5.2 | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 21.0 Å | |||||||||
 Authors Authors | Cottrell CA / Ward AB | |||||||||
 Citation Citation |  Journal: Cell / Year: 2019 Journal: Cell / Year: 2019Title: Slow Delivery Immunization Enhances HIV Neutralizing Antibody and Germinal Center Responses via Modulation of Immunodominance. Authors: Kimberly M Cirelli / Diane G Carnathan / Bartek Nogal / Jacob T Martin / Oscar L Rodriguez / Amit A Upadhyay / Chiamaka A Enemuo / Etse H Gebru / Yury Choe / Federico Viviano / Catherine ...Authors: Kimberly M Cirelli / Diane G Carnathan / Bartek Nogal / Jacob T Martin / Oscar L Rodriguez / Amit A Upadhyay / Chiamaka A Enemuo / Etse H Gebru / Yury Choe / Federico Viviano / Catherine Nakao / Matthias G Pauthner / Samantha Reiss / Christopher A Cottrell / Melissa L Smith / Raiza Bastidas / William Gibson / Amber N Wolabaugh / Mariane B Melo / Benjamin Cossette / Venkatesh Kumar / Nirav B Patel / Talar Tokatlian / Sergey Menis / Daniel W Kulp / Dennis R Burton / Ben Murrell / William R Schief / Steven E Bosinger / Andrew B Ward / Corey T Watson / Guido Silvestri / Darrell J Irvine / Shane Crotty /   Abstract: Conventional immunization strategies will likely be insufficient for the development of a broadly neutralizing antibody (bnAb) vaccine for HIV or other difficult pathogens because of the ...Conventional immunization strategies will likely be insufficient for the development of a broadly neutralizing antibody (bnAb) vaccine for HIV or other difficult pathogens because of the immunological hurdles posed, including B cell immunodominance and germinal center (GC) quantity and quality. We found that two independent methods of slow delivery immunization of rhesus monkeys (RMs) resulted in more robust T follicular helper (T) cell responses and GC B cells with improved Env-binding, tracked by longitudinal fine needle aspirates. Improved GCs correlated with the development of >20-fold higher titers of autologous nAbs. Using a new RM genomic immunoglobulin locus reference, we identified differential IgV gene use between immunization modalities. Ab mapping demonstrated targeting of immunodominant non-neutralizing epitopes by conventional bolus-immunized animals, whereas slow delivery-immunized animals targeted a more diverse set of epitopes. Thus, alternative immunization strategies can enhance nAb development by altering GCs and modulating the immunodominance of non-neutralizing epitopes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9138.map.gz emd_9138.map.gz | 20.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9138-v30.xml emd-9138-v30.xml emd-9138.xml emd-9138.xml | 16 KB 16 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9138.png emd_9138.png | 25.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9138 http://ftp.pdbj.org/pub/emdb/structures/EMD-9138 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9138 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9138 | HTTPS FTP |
-Related structure data
| Related structure data | 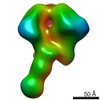 0569C 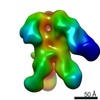 0570C  0571C 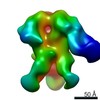 0572C  0573C  0574C 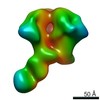 0575C  0576C  0577C 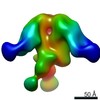 0578C  0579C  0580C  0581C  0582C 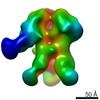 9175C 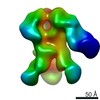 9176C  9177C  9178C  9179C  9180C  9181C  9182C  9183C  9184C  9185C  9186C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9138.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9138.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | HIV-1 vaccine elicited Fab BDA1 in complex with the HIV Env trimer BG505 SOSIPv5.2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.98 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
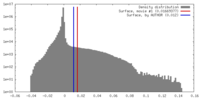
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : HIV-1 vaccine elicited Fab BDA1 in complex with the HIV Env trime...
| Entire | Name: HIV-1 vaccine elicited Fab BDA1 in complex with the HIV Env trimer BG505 SOSIPv5.2 |
|---|---|
| Components |
|
-Supramolecule #1: HIV-1 vaccine elicited Fab BDA1 in complex with the HIV Env trime...
| Supramolecule | Name: HIV-1 vaccine elicited Fab BDA1 in complex with the HIV Env trimer BG505 SOSIPv5.2 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: SOSIP trimer
| Supramolecule | Name: SOSIP trimer / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Supramolecule #3: BDA1 Fab
| Supramolecule | Name: BDA1 Fab / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: BG505 SOSIPv5.2 gp120
| Macromolecule | Name: BG505 SOSIPv5.2 gp120 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AENLWVTVYY GVPVWKDAET TLFCASDAKA YETKKHNVWA THCCVPTDPN PQEIHLENVT EEFNMWKNNM VEQMHTDIIS LWDQSLKPCV KLTPLCVTLQ CTNVTNNITD DMRGELKNCS FNMTTELRDK KQKVYSLFYR LDVVQINENQ GNRSNNSNKE YRLINCNTSA ...String: AENLWVTVYY GVPVWKDAET TLFCASDAKA YETKKHNVWA THCCVPTDPN PQEIHLENVT EEFNMWKNNM VEQMHTDIIS LWDQSLKPCV KLTPLCVTLQ CTNVTNNITD DMRGELKNCS FNMTTELRDK KQKVYSLFYR LDVVQINENQ GNRSNNSNKE YRLINCNTSA ITQACPKVSF EPIPIHYCAP AGFAILKCKD KKFNGTGPCP SVSTVQCTHG IKPVVSTQLL LNGSLAEEEV MIRSENITNN AKNILVQFNT PVQINCTRPN NNTRKSIRIG PGQWFYATGD IIGDIRQAHC NVSKATWNET LGKVVKQLRK HFGNNTIIRF ANSSGGDLEV TTHSFNCGGE FFYCNTSGLF NSTWISNTSV QGSNSTGSND SITLPCRIKQ IINMWQRIGQ AMYAPPIQGV IRCVSNITGL ILTRDGGSTN STTETFRPGG GDMRDNWRSE LYKYKVVKIE PLGVAPTRCK RRVVGRRRRR R |
-Macromolecule #2: BG505 SOSIPv5.2 gp41
| Macromolecule | Name: BG505 SOSIPv5.2 gp41 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVGIGAVFLG FLGAAGSTMG AASMTLTVQA RNLLSGIVQQ QSNLLRAPEC QQHLLKLTVW GIKQLQARVL AVERYLRDQQ LLGIWGCSGK LICCTNVPWN SSWSNRNLSE IWDNMTWLQW DKEISNYTQI IYGLLEESQN QQEKNEQDLL ALD |
-Macromolecule #3: BDA1 Fab heavy chain
| Macromolecule | Name: BDA1 Fab heavy chain / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLQESGPG LVKPSETLSL TCAVSGASIS IYWWGWIRQP PGKGLEWIGE IIGSSGSTNS NPSFKSRVTI SKDASKNQFS LNLNSVTAAD TAVYYCVRVG AAISLPFDYW GQGVLVTVSS |
-Macromolecule #4: BDA1 Fab light chain
| Macromolecule | Name: BDA1 Fab light chain / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SYELTQPPSV SVSPGQTARI TCSGDALPKK YAYWFQQKPG QSPVLIIYED NKRPSGIPER FSGSSSGTVA TLTISGAQVE DEGDYYCYSR HSSGNHGLFG GGTRLTVL |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl Formate |
| Grid | Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI CETA (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Sampling interval: 14.0 µm / Number grids imaged: 1 / Average exposure time: 1.0 sec. / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



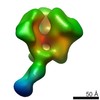





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)