+Search query
-Structure paper
| Title | Slow Delivery Immunization Enhances HIV Neutralizing Antibody and Germinal Center Responses via Modulation of Immunodominance. |
|---|---|
| Journal, issue, pages | Cell, Vol. 177, Issue 5, Page 1153-1171.e28, Year 2019 |
| Publish date | May 16, 2019 |
 Authors Authors | Kimberly M Cirelli / Diane G Carnathan / Bartek Nogal / Jacob T Martin / Oscar L Rodriguez / Amit A Upadhyay / Chiamaka A Enemuo / Etse H Gebru / Yury Choe / Federico Viviano / Catherine Nakao / Matthias G Pauthner / Samantha Reiss / Christopher A Cottrell / Melissa L Smith / Raiza Bastidas / William Gibson / Amber N Wolabaugh / Mariane B Melo / Benjamin Cossette / Venkatesh Kumar / Nirav B Patel / Talar Tokatlian / Sergey Menis / Daniel W Kulp / Dennis R Burton / Ben Murrell / William R Schief / Steven E Bosinger / Andrew B Ward / Corey T Watson / Guido Silvestri / Darrell J Irvine / Shane Crotty /   |
| PubMed Abstract | Conventional immunization strategies will likely be insufficient for the development of a broadly neutralizing antibody (bnAb) vaccine for HIV or other difficult pathogens because of the ...Conventional immunization strategies will likely be insufficient for the development of a broadly neutralizing antibody (bnAb) vaccine for HIV or other difficult pathogens because of the immunological hurdles posed, including B cell immunodominance and germinal center (GC) quantity and quality. We found that two independent methods of slow delivery immunization of rhesus monkeys (RMs) resulted in more robust T follicular helper (T) cell responses and GC B cells with improved Env-binding, tracked by longitudinal fine needle aspirates. Improved GCs correlated with the development of >20-fold higher titers of autologous nAbs. Using a new RM genomic immunoglobulin locus reference, we identified differential IgV gene use between immunization modalities. Ab mapping demonstrated targeting of immunodominant non-neutralizing epitopes by conventional bolus-immunized animals, whereas slow delivery-immunized animals targeted a more diverse set of epitopes. Thus, alternative immunization strategies can enhance nAb development by altering GCs and modulating the immunodominance of non-neutralizing epitopes. |
 External links External links |  Cell / Cell /  PubMed:31080066 / PubMed:31080066 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 16.4 - 29.5 Å |
| Structure data |  EMDB-0569:  EMDB-0570:  EMDB-0571:  EMDB-0572:  EMDB-0573:  EMDB-0574:  EMDB-0575:  EMDB-0576:  EMDB-0577:  EMDB-0578:  EMDB-0579:  EMDB-0580:  EMDB-0581:  EMDB-0582:  EMDB-9138:  EMDB-9175:  EMDB-9176: 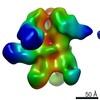 EMDB-9177:  EMDB-9178:  EMDB-9179:  EMDB-9180: 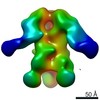 EMDB-9181: 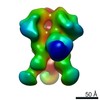 EMDB-9182: 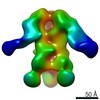 EMDB-9183:  EMDB-9184:  EMDB-9185:  EMDB-9186: |
| Source |
|
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers



 Homo sapiens (human)
Homo sapiens (human)

 Human immunodeficiency virus 1
Human immunodeficiency virus 1