+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21449 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CON-S ch.DS.SOSIP HIV trimer bound to DH842 Fab | ||||||||||||
 Map data Map data | CON-S ch.DS.SOSIP HIV trimer bound to DH842 Fab | ||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | ||||||||||||
| Method | single particle reconstruction / negative staining / Resolution: 17.0 Å | ||||||||||||
 Authors Authors | Cai F / Chen WH / Wu W / Jones J / Misook C / Gohain N / Shen X / LaBranche C / Eaton A / Sutherland L ...Cai F / Chen WH / Wu W / Jones J / Misook C / Gohain N / Shen X / LaBranche C / Eaton A / Sutherland L / Hernandez G / Nelson RW / Scearce R / Seaman M / Moody MA / Santra S / Wiehe KJ / Tomaras GD / Wagh K / Korber B / Bonsignori M / Montefiori DC / Haynes BF / de Val N / Joyce GM / Saunders KO | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: HIV-1 vaccination elicits structurally and genetically-convergent neutralizing antibodies in non-human primates Authors: Cai F / Chen WH / Wu W / Jones J / Misook C / Gohain N / Shen X / LaBranche C / Eaton A / Sutherland L / Hernandez G / Nelson RW / Scearce R / Seaman M / Moody MA / Santra S / Wiehe KJ / ...Authors: Cai F / Chen WH / Wu W / Jones J / Misook C / Gohain N / Shen X / LaBranche C / Eaton A / Sutherland L / Hernandez G / Nelson RW / Scearce R / Seaman M / Moody MA / Santra S / Wiehe KJ / Tomaras GD / Wagh K / Korber B / Bonsignori M / Montefiori DC / Haynes BF / de Val N / Joyce GM / Saunders KO | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21449.map.gz emd_21449.map.gz | 2.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21449-v30.xml emd-21449-v30.xml emd-21449.xml emd-21449.xml | 12.5 KB 12.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_21449.png emd_21449.png | 20.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21449 http://ftp.pdbj.org/pub/emdb/structures/EMD-21449 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21449 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21449 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_21449.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21449.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
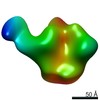
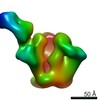
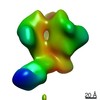
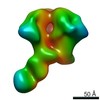
| Annotation | CON-S ch.DS.SOSIP HIV trimer bound to DH842 Fab | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.19 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : CON-S ch.DS.SOSIP in complex with DH842 Fab
| Entire | Name: CON-S ch.DS.SOSIP in complex with DH842 Fab |
|---|---|
| Components |
|
-Supramolecule #1: CON-S ch.DS.SOSIP in complex with DH842 Fab
| Supramolecule | Name: CON-S ch.DS.SOSIP in complex with DH842 Fab / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK 293F cells Homo sapiens (human) / Recombinant cell: HEK 293F cells |
| Molecular weight | Experimental: 50 KDa |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Staining | Type: NEGATIVE / Material: Uranyl Formate Details: grids were glow discharged and using 0.7% UF they were stained. | |||||||||
| Grid | Model: C-flat / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 20 |
|---|---|
| Image recording | Film or detector model: FEI EAGLE (2k x 2k) / Number grids imaged: 1 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.1 µm / Nominal defocus min: 1.1 µm / Nominal magnification: 100000 |
| Sample stage | Specimen holder model: OTHER |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera





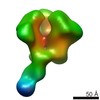
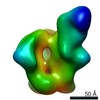
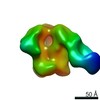
 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















