[English] 日本語
 Yorodumi
Yorodumi- EMDB-9038: B41 SOSIP.664 in complex with soluble CD4 (D1-D2), the co-recepto... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9038 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | B41 SOSIP.664 in complex with soluble CD4 (D1-D2), the co-receptor mimicking antibody 21c and the broadly neutralizing antibody 8ANC195 | |||||||||
 Map data Map data | Sharpened map of complex used for model building and refinement. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HIV-1 Env / broadly neutralizing antibodies / cryo-EM / single particle analysis / VIRAL PROTEIN-IMMUNE SYSTEM complex / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationhelper T cell enhancement of adaptive immune response / interleukin-16 binding / interleukin-16 receptor activity / maintenance of protein location in cell / response to methamphetamine hydrochloride / cellular response to ionomycin / T cell selection / MHC class II protein binding / positive regulation of kinase activity / cellular response to granulocyte macrophage colony-stimulating factor stimulus ...helper T cell enhancement of adaptive immune response / interleukin-16 binding / interleukin-16 receptor activity / maintenance of protein location in cell / response to methamphetamine hydrochloride / cellular response to ionomycin / T cell selection / MHC class II protein binding / positive regulation of kinase activity / cellular response to granulocyte macrophage colony-stimulating factor stimulus / interleukin-15-mediated signaling pathway / positive regulation of monocyte differentiation / Alpha-defensins / Nef Mediated CD4 Down-regulation / regulation of T cell activation / response to vitamin D / extracellular matrix structural constituent / Other interleukin signaling / T cell receptor complex / enzyme-linked receptor protein signaling pathway / Translocation of ZAP-70 to Immunological synapse / Phosphorylation of CD3 and TCR zeta chains / positive regulation of protein kinase activity / regulation of calcium ion transport / macrophage differentiation / Generation of second messenger molecules / positive regulation of calcium ion transport into cytosol / immunoglobulin binding / T cell differentiation / Co-inhibition by PD-1 / Binding and entry of HIV virion / symbiont-mediated perturbation of host defense response / coreceptor activity / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of calcium-mediated signaling / positive regulation of interleukin-2 production / positive regulation of T cell proliferation / cell surface receptor protein tyrosine kinase signaling pathway / protein tyrosine kinase binding / host cell endosome membrane / Vpu mediated degradation of CD4 / clathrin-coated endocytic vesicle membrane / calcium-mediated signaling / positive regulation of protein phosphorylation / transmembrane signaling receptor activity / MHC class II protein complex binding / response to estradiol / Downstream TCR signaling / Cargo recognition for clathrin-mediated endocytosis / signaling receptor activity / Clathrin-mediated endocytosis / virus receptor activity / clathrin-dependent endocytosis of virus by host cell / response to ethanol / defense response to Gram-negative bacterium / adaptive immune response / positive regulation of viral entry into host cell / early endosome / cell surface receptor signaling pathway / positive regulation of canonical NF-kappaB signal transduction / positive regulation of ERK1 and ERK2 cascade / cell adhesion / positive regulation of MAPK cascade / immune response / viral protein processing / membrane raft / endoplasmic reticulum lumen / fusion of virus membrane with host plasma membrane / external side of plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / lipid binding / protein kinase binding / endoplasmic reticulum membrane / positive regulation of DNA-templated transcription / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / enzyme binding / signal transduction / protein homodimerization activity / zinc ion binding / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||
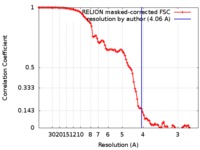
| Method | single particle reconstruction / cryo EM / Resolution: 4.06 Å | |||||||||
 Authors Authors | Barnes CO / Bjorkman PJ | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Cell Host Microbe / Year: 2018 Journal: Cell Host Microbe / Year: 2018Title: Partially Open HIV-1 Envelope Structures Exhibit Conformational Changes Relevant for Coreceptor Binding and Fusion. Authors: Haoqing Wang / Christopher O Barnes / Zhi Yang / Michel C Nussenzweig / Pamela J Bjorkman /  Abstract: HIV-1 Env, a trimer of gp120-gp41 heterodimers, mediates membrane fusion after binding host receptor CD4. Receptor binding displaces V1V2 loops from Env's apex, allowing coreceptor binding and ...HIV-1 Env, a trimer of gp120-gp41 heterodimers, mediates membrane fusion after binding host receptor CD4. Receptor binding displaces V1V2 loops from Env's apex, allowing coreceptor binding and opening Env to enable gp41-mediated fusion. We present 3.54 Å and 4.06 Å cryoelectron microscopy structures of partially open soluble native-like Env trimers (SOSIPs) bound to CD4. One structure, a complex with a coreceptor-mimicking antibody that binds both CD4 and gp120, stabilizes the displaced V1V2 and reveals its structure. Comparing partially and fully open Envs with closed Envs shows that gp41 rearrangements are independent of the CD4-induced rearrangements that result in V1V2 displacement and formation of a 4-stranded bridging sheet. These findings suggest ordered conformational changes before coreceptor binding: (1) gp120 opening inducing side-chain rearrangements and a compact gp41 central helix conformation, and (2) 4-stranded bridging-sheet formation and V1V2 displacement. These analyses illuminate potential receptor-induced Env changes and inform design of therapeutics disrupting viral entry. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9038.map.gz emd_9038.map.gz | 85.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9038-v30.xml emd-9038-v30.xml emd-9038.xml emd-9038.xml | 24 KB 24 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9038_fsc.xml emd_9038_fsc.xml | 10.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_9038.png emd_9038.png | 96.9 KB | ||
| Masks |  emd_9038_msk_1.map emd_9038_msk_1.map emd_9038_msk_2.map emd_9038_msk_2.map | 91.1 MB 91.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-9038.cif.gz emd-9038.cif.gz | 8.1 KB | ||
| Others |  emd_9038_additional.map.gz emd_9038_additional.map.gz | 85.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9038 http://ftp.pdbj.org/pub/emdb/structures/EMD-9038 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9038 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9038 | HTTPS FTP |
-Related structure data
| Related structure data |  6eduMC  7516C  6cm3C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9038.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9038.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map of complex used for model building and refinement. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.31 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_9038_msk_1.map emd_9038_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Mask #2
| File |  emd_9038_msk_2.map emd_9038_msk_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Sharpened map of focused refinement of 21c, gp120,...
| File | emd_9038_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map of focused refinement of 21c, gp120, and sCD4. Used for model building and refinement of V1V2 region, 21c-gp120 interface, and 21c-sCD4 interface. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : B41 SOSIP-sCD4-21c-8ANC195 complex
| Entire | Name: B41 SOSIP-sCD4-21c-8ANC195 complex |
|---|---|
| Components |
|
-Supramolecule #1: B41 SOSIP-sCD4-21c-8ANC195 complex
| Supramolecule | Name: B41 SOSIP-sCD4-21c-8ANC195 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#7 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 700 KDa |
-Macromolecule #1: Envelope glycoprotein gp160
| Macromolecule | Name: Envelope glycoprotein gp160 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 17.357824 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AVGLGAFILG FLGAAGSTMG AASMALTVQA RLLLSGIVQQ QNNLLRAPEA QQHMLQLTVW GIKQLQARVL AVERYLRDQQ LLGIWGCSG KIICCTNVPW NDSWSNKTIN EIWDNMTWMQ WEKEIDNYTQ HIYTLLEVSQ IQQEKNEQEL LELD UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #2: Envelope glycoprotein gp160
| Macromolecule | Name: Envelope glycoprotein gp160 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 57.702469 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDAMKRGLCC VLLLCGAVFV SPSQEIHARF RRGARAAKKW VTVYYGVPVW KEATTTLFCA SDAKAYDTEV HNVWATHACV PTDPNPQEI VLGNVTENFN MWKNNMVEQM HEDIISLWDQ SLKPCVKLTP LCVTLNCNNV NTNNTNNSTN ATISDWEKME T GEMKNCSF ...String: MDAMKRGLCC VLLLCGAVFV SPSQEIHARF RRGARAAKKW VTVYYGVPVW KEATTTLFCA SDAKAYDTEV HNVWATHACV PTDPNPQEI VLGNVTENFN MWKNNMVEQM HEDIISLWDQ SLKPCVKLTP LCVTLNCNNV NTNNTNNSTN ATISDWEKME T GEMKNCSF NVTTSIRDKI KKEYALFYKL DVVPLENKNN INNTNITNYR LINCNTSVIT QACPKVSFEP IPIHYCAPAG FA ILKCNSK TFNGSGPCTN VSTVQCTHGI RPVVSTQLLL NGSLAEEEIV IRSENITDNA KTIIVQLNEA VEINCTRPNN NTR KSIHIG PGRAFYATGD IIGNIRQAHC NISKARWNET LGQIVAKLEE QFPNKTIIFN HSSGGDPEIV THSFNCGGEF FYCN TTPLF NSTWNNTRTD DYPTGGEQNI TLQCRIKQII NMWQGVGKAM YAPPIRGQIR CSSNITGLLL TRDGGRDQNG TETFR PGGG NMRDNWRSEL YKYKVVKIEP LGIAPTACKR RV UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #3: T-cell surface glycoprotein CD4
| Macromolecule | Name: T-cell surface glycoprotein CD4 / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 20.50326 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: KKVVLGKKGD TVELTCTASQ KKSIQFHWKN SNQIKILGNQ GSFLTKGPSK LNDRADSRRS LWDQGNFPLI IKNLKIEDSD TYICEVEDQ KEEVQLLVFG LTANSDTHLL QGQSLTLTLE SPPGSSPSVQ CRSPRGKNIQ GGKTLSVSQL ELQDSGTWTC T VLQNQKKV EFKIDIVVLA FQKASNT UniProtKB: T-cell surface glycoprotein CD4 |
-Macromolecule #4: 21c Fab VH domain
| Macromolecule | Name: 21c Fab VH domain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.61959 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQVVQSGAE VRKPGASVKV SCKVSGFTLT GLSIHWVRQA PGKGLEWMGG FGPEENEIIY AQKFQGRVSM TEDTSTNTAY MELSSLRSE DTAVYYCATG GNYYNLWTGY YPLAYWGQGT LVTVSSASTK GPSVFPLAPS SKSTSGGTAA LGCLVKDYFP E PVTVSWNS ...String: QVQVVQSGAE VRKPGASVKV SCKVSGFTLT GLSIHWVRQA PGKGLEWMGG FGPEENEIIY AQKFQGRVSM TEDTSTNTAY MELSSLRSE DTAVYYCATG GNYYNLWTGY YPLAYWGQGT LVTVSSASTK GPSVFPLAPS SKSTSGGTAA LGCLVKDYFP E PVTVSWNS GALTSGVHTF PAVLQSSGLY SLSSVVTVPS SSLGTQTYIC NVNHKPSNTK VDKKVEPKSC DKT |
-Macromolecule #5: 21c Fab VL domain
| Macromolecule | Name: 21c Fab VL domain / type: protein_or_peptide / ID: 5 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.815264 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSVLTQPPSV SAAPGQKVTI SCSGSSSNIG KNYVSWYQQL PGAAPKLLIF DDTQRPSGIP DRFSGSKSGT SATLAITGLQ TGDEADYYC GTWDSSLSTG QLFGGGTKLT VLGQPKAAPS VTLFPPSSEE LQANKATLVC LISDFYPGAV TVAWKADSSP V KAGVETTT ...String: QSVLTQPPSV SAAPGQKVTI SCSGSSSNIG KNYVSWYQQL PGAAPKLLIF DDTQRPSGIP DRFSGSKSGT SATLAITGLQ TGDEADYYC GTWDSSLSTG QLFGGGTKLT VLGQPKAAPS VTLFPPSSEE LQANKATLVC LISDFYPGAV TVAWKADSSP V KAGVETTT PSKQSNNKYA ASSYLSLTPE QWKSHRSYSC QVTHEGSTVE KTMAHAECS |
-Macromolecule #6: 8ANC195 G52K5 Fab VH domain
| Macromolecule | Name: 8ANC195 G52K5 Fab VH domain / type: protein_or_peptide / ID: 6 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 26.12527 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QIHLVQSGTE VKKPGSSVTV SCKAYGVNTF GLYAVNWVRQ APGQSLEYIG QIWRWKSSAS HHFRGRVLIS AVDLTGSSPP ISSLEIKNL TSDDTAVYFC TTTSTYDKWS GLHHDGVMAF SSWGQGTLIS VSAASTKGPS VFPLAPSSKS TSGGTAALGC L VKDYFPEP ...String: QIHLVQSGTE VKKPGSSVTV SCKAYGVNTF GLYAVNWVRQ APGQSLEYIG QIWRWKSSAS HHFRGRVLIS AVDLTGSSPP ISSLEIKNL TSDDTAVYFC TTTSTYDKWS GLHHDGVMAF SSWGQGTLIS VSAASTKGPS VFPLAPSSKS TSGGTAALGC L VKDYFPEP VTVSWNSGAL TSGVHTFPAV LQSSGLYSLS SVVTVPSSSL GTQTYICNVN HKPSNTKVDK RVEPKSCDKT HH HHHH |
-Macromolecule #7: 8ANC195 G52K5 Fab VL domain
| Macromolecule | Name: 8ANC195 G52K5 Fab VL domain / type: protein_or_peptide / ID: 7 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.401984 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPST LSASIGDTVR ISCRASQSIT GNWVAWYQQR PGKAPRLLIY RGAALLGGVP SRFSGSAAGT DFTLTIGNLQ AEDFGTFYC QQYDTYPGTF GQGTKVEVKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: DIQMTQSPST LSASIGDTVR ISCRASQSIT GNWVAWYQQR PGKAPRLLIY RGAALLGGVP SRFSGSAAGT DFTLTIGNLQ AEDFGTFYC QQYDTYPGTF GQGTKVEVKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Macromolecule #13: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 13 / Number of copies: 27 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.35 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Details: 0.26 mA | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 1 / Number real images: 2531 / Average exposure time: 10.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.7 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Correlation coefficient |
|---|---|
| Output model |  PDB-6edu: |
 Movie
Movie Controller
Controller





























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)