[English] 日本語
 Yorodumi
Yorodumi- EMDB-8865: Negative-stain reconstruction of an N-terminally MBP tagged Poz1 ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8865 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Negative-stain reconstruction of an N-terminally MBP tagged Poz1 in the S. pombe CTP complex (Ccq1 2-716, Tpz1 406-508, Poz1 2-249) | |||||||||
 Map data Map data | Negative-stain reconstruction of an N-terminally MBP tagged Poz1 in the S. pombe CTP complex (Ccq1 2-716, Tpz1 406-508, Poz1 2-249) | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationtelomere-telomerase complex assembly / subtelomeric heterochromatin / telomere cap complex / chromosome, telomeric repeat region / meiotic chromosome segregation / meiotic telomere clustering / telomere maintenance via telomere lengthening / shelterin complex / telomere capping / telomerase holoenzyme complex ...telomere-telomerase complex assembly / subtelomeric heterochromatin / telomere cap complex / chromosome, telomeric repeat region / meiotic chromosome segregation / meiotic telomere clustering / telomere maintenance via telomere lengthening / shelterin complex / telomere capping / telomerase holoenzyme complex / protein localization to chromosome, telomeric region / telomeric DNA binding / telomere maintenance via telomerase / telomere organization / telomere maintenance / molecular adaptor activity / DNA binding / nucleoplasm / nucleus / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
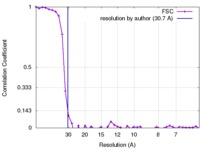
| Method | single particle reconstruction / negative staining / Resolution: 30.7 Å | |||||||||
 Authors Authors | Scott HW / Kim JK / Yu C / Huang L / Qiao F / Taylor DJ | |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2017 Journal: J Mol Biol / Year: 2017Title: Spatial Organization and Molecular Interactions of the Schizosaccharomyces pombe Ccq1-Tpz1-Poz1 Shelterin Complex. Authors: Harry Scott / Jin-Kwang Kim / Clinton Yu / Lan Huang / Feng Qiao / Derek J Taylor /  Abstract: The shelterin complex is a macromolecular assembly of proteins that binds to and protects telomeric DNA, which composes the ends of all linear chromosomes. Shelterin proteins prevent chromosome ends ...The shelterin complex is a macromolecular assembly of proteins that binds to and protects telomeric DNA, which composes the ends of all linear chromosomes. Shelterin proteins prevent chromosome ends from fusing together and from eliciting erroneous induction of DNA damage response pathways. In addition, shelterin proteins play key roles in regulating the recruitment and activation of telomerase, an enzyme that extends telomeric DNA. In fission yeast, Schizosaccharomyces pombe, interactions between the shelterin proteins Ccq1, Tpz1, and Poz1 are important for regulating telomerase-mediated telomere synthesis and thus telomere length homeostasis. Here, we used electron microscopy combined with genetic labeling to define the three-dimensional arrangement of the S. pombe Ccq1-Tpz1-Poz1 (CTP) complex. Crosslinking mass spectrometry was used to identify individual residues that are in proximity to the protein-protein interfaces of the assembled CTP complex. Together, our data provide a first glimpse into the architectural design of the CTP complex and reveals unique interactions that are important in maintaining the S. pombe telomere in a non-extendible state. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8865.map.gz emd_8865.map.gz | 2.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8865-v30.xml emd-8865-v30.xml emd-8865.xml emd-8865.xml | 12.4 KB 12.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8865_fsc.xml emd_8865_fsc.xml | 4.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_8865.png emd_8865.png | 11.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8865 http://ftp.pdbj.org/pub/emdb/structures/EMD-8865 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8865 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8865 | HTTPS FTP |
-Related structure data
| Related structure data |  8863C  8864C  8866C  8861 C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8865.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8865.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative-stain reconstruction of an N-terminally MBP tagged Poz1 in the S. pombe CTP complex (Ccq1 2-716, Tpz1 406-508, Poz1 2-249) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.004 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Negative-stain reconstruction of an N-terminally MBP tagged Poz1 ...
| Entire | Name: Negative-stain reconstruction of an N-terminally MBP tagged Poz1 in the S. pombe CTP complex (Ccq1 2-716, Tpz1 406-508, Poz1 2-249) |
|---|---|
| Components |
|
-Supramolecule #1: Negative-stain reconstruction of an N-terminally MBP tagged Poz1 ...
| Supramolecule | Name: Negative-stain reconstruction of an N-terminally MBP tagged Poz1 in the S. pombe CTP complex (Ccq1 2-716, Tpz1 406-508, Poz1 2-249) type: complex / ID: 1 / Parent: 0 / Details: Complex is a dimer of trimers |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.064 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Staining | Type: NEGATIVE / Material: Uranyl acetate | ||||||||||||
| Grid | Pretreatment - Type: GLOW DISCHARGE / Details: 15 mA |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2200FS |
|---|---|
| Specialist optics | Energy filter - Name: omega in-column |
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Digitization - Dimensions - Width: 4000 pixel / Digitization - Dimensions - Height: 4000 pixel / Digitization - Sampling interval: 15.6 µm / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)