+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8827 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cas1-Cas2-IHF-DNA holo-complex | |||||||||
 Map data Map data | Cas1-Cas2-IHF-DNA holo-complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR integration complex / DNA / Cas1-Cas2 / IHF / DNA BINDING PROTEIN / DNA BINDING PROTEIN-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of metabolic process / CRISPR-cas system / crossover junction DNA endonuclease activity / 5'-flap endonuclease activity / maintenance of CRISPR repeat elements / structural constituent of chromatin / regulation of translation / chromosome / endonuclease activity / DNA recombination ...positive regulation of metabolic process / CRISPR-cas system / crossover junction DNA endonuclease activity / 5'-flap endonuclease activity / maintenance of CRISPR repeat elements / structural constituent of chromatin / regulation of translation / chromosome / endonuclease activity / DNA recombination / defense response to virus / Hydrolases; Acting on ester bonds / DNA repair / DNA damage response / regulation of DNA-templated transcription / protein homodimerization activity / DNA binding / metal ion binding / identical protein binding / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |    | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.64 Å | |||||||||
 Authors Authors | Wright AV / Liu JJ | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2017 Journal: Science / Year: 2017Title: Structures of the CRISPR genome integration complex. Authors: Addison V Wright / Jun-Jie Liu / Gavin J Knott / Kevin W Doxzen / Eva Nogales / Jennifer A Doudna /  Abstract: CRISPR-Cas systems depend on the Cas1-Cas2 integrase to capture and integrate short foreign DNA fragments into the CRISPR locus, enabling adaptation to new viruses. We present crystal structures of ...CRISPR-Cas systems depend on the Cas1-Cas2 integrase to capture and integrate short foreign DNA fragments into the CRISPR locus, enabling adaptation to new viruses. We present crystal structures of Cas1-Cas2 bound to both donor and target DNA in intermediate and product integration complexes, as well as a cryo-electron microscopy structure of the full CRISPR locus integration complex, including the accessory protein IHF (integration host factor). The structures show unexpectedly that indirect sequence recognition dictates integration site selection by favoring deformation of the repeat and the flanking sequences. IHF binding bends the DNA sharply, bringing an upstream recognition motif into contact with Cas1 to increase both the specificity and efficiency of integration. These results explain how the Cas1-Cas2 CRISPR integrase recognizes a sequence-dependent DNA structure to ensure site-selective CRISPR array expansion during the initial step of bacterial adaptive immunity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8827.map.gz emd_8827.map.gz | 62.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8827-v30.xml emd-8827-v30.xml emd-8827.xml emd-8827.xml | 20.4 KB 20.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8827.png emd_8827.png | 99.9 KB | ||
| Filedesc metadata |  emd-8827.cif.gz emd-8827.cif.gz | 6.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8827 http://ftp.pdbj.org/pub/emdb/structures/EMD-8827 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8827 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8827 | HTTPS FTP |
-Related structure data
| Related structure data |  5wfeMC  5vvjC  5vvkC  5vvlC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8827.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8827.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cas1-Cas2-IHF-DNA holo-complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
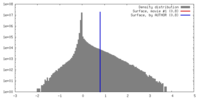
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Cas1-Cas2-IHF-DNA holo complex
| Entire | Name: Cas1-Cas2-IHF-DNA holo complex |
|---|---|
| Components |
|
-Supramolecule #1: Cas1-Cas2-IHF-DNA holo complex
| Supramolecule | Name: Cas1-Cas2-IHF-DNA holo complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#8 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: CRISPR-associated endonuclease Cas1
| Macromolecule | Name: CRISPR-associated endonuclease Cas1 / type: protein_or_peptide / ID: 1 / Details: Cas1 / Number of copies: 4 / Enantiomer: LEVO / EC number: Hydrolases; Acting on ester bonds |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 33.235418 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTWLPLNPIP LKDRVSMIFL QYGQIDVIDG AFVLIDKTGI RTHIPVGSVA CIMLEPGTRV SHAAVRLAAQ VGTLLVWVGE AGVRVYASG QPGGARSDKL LYQAKLALDE DLRLKVVRKM FELRFGEPAP ARRSVEQLRG IEGSRVRATY ALLAKQYGVT W NGRRYDPK ...String: MTWLPLNPIP LKDRVSMIFL QYGQIDVIDG AFVLIDKTGI RTHIPVGSVA CIMLEPGTRV SHAAVRLAAQ VGTLLVWVGE AGVRVYASG QPGGARSDKL LYQAKLALDE DLRLKVVRKM FELRFGEPAP ARRSVEQLRG IEGSRVRATY ALLAKQYGVT W NGRRYDPK DWEKGDTINQ CISAATSCLY GVTEAAILAA GYAPAIGFVH TGKPLSFVYD IADIIKFDTV VPKAFEIARR NP GEPDREV RLACRDIFRS SKTLAKLIPL IEDVLAAGEI QPPAPPEDAQ PVAIPLPVSL GDAGHRSS UniProtKB: CRISPR-associated endonuclease Cas1 |
-Macromolecule #2: CRISPR-associated endoribonuclease Cas2
| Macromolecule | Name: CRISPR-associated endoribonuclease Cas2 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO / EC number: Hydrolases; Acting on ester bonds |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.55327 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSMLVVVTEN VPPRLRGRLA IWLLEVRAGV YVGDVSAKIR EMIWEQIAGL AEEGNVVMAW ATNTETGFEF QTFGLNRRTP VDLDGLRLV SFLPVGSSEN LYFQ UniProtKB: CRISPR-associated endoribonuclease Cas2 |
-Macromolecule #7: Integration host factor subunit alpha
| Macromolecule | Name: Integration host factor subunit alpha / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.373952 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MALTKAEMSE YLFDKLGLSK RDAKELVELF FEEIRRALEN GEQVKLSGFG NFDLRDKNQR PGRNPKTGED IPITARRVVT FRPGQKLKS RVENASPKDE UniProtKB: Integration host factor subunit alpha |
-Macromolecule #8: Integration host factor subunit beta
| Macromolecule | Name: Integration host factor subunit beta / type: protein_or_peptide / ID: 8 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 10.671178 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTKSELIERL ATQQSHIPAK TVEDAVKEML EHMASTLAQG ERIEIRGFGS FSLHYRAPRT GRNPKTGDKV ELEGKYVPHF KPGKELRDR ANIYG UniProtKB: Integration host factor subunit beta |
-Macromolecule #3: DNA (28-MER)
| Macromolecule | Name: DNA (28-MER) / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 8.680646 KDa |
| Sequence | String: (DA)(DA)(DA)(DC)(DA)(DC)(DC)(DA)(DG)(DA) (DA)(DC)(DG)(DA)(DG)(DT)(DA)(DG)(DT)(DA) (DA)(DA)(DT)(DT)(DG)(DG)(DG)(DC) |
-Macromolecule #4: DNA (45-MER)
| Macromolecule | Name: DNA (45-MER) / type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 18.745965 KDa |
| Sequence | String: (DA)(DT)(DT)(DT)(DA)(DC)(DT)(DA)(DC)(DT) (DC)(DG)(DT)(DT)(DC)(DT)(DG)(DG)(DT)(DG) (DT)(DT)(DT)(DC)(DT)(DC)(DG)(DT)(DG) (DT)(DG)(DT)(DT)(DC)(DC)(DC)(DC)(DG)(DC) (DG) (DC)(DC)(DA)(DG)(DC)(DG) ...String: (DA)(DT)(DT)(DT)(DA)(DC)(DT)(DA)(DC)(DT) (DC)(DG)(DT)(DT)(DC)(DT)(DG)(DG)(DT)(DG) (DT)(DT)(DT)(DC)(DT)(DC)(DG)(DT)(DG) (DT)(DG)(DT)(DT)(DC)(DC)(DC)(DC)(DG)(DC) (DG) (DC)(DC)(DA)(DG)(DC)(DG)(DG)(DG) (DG)(DA)(DT)(DA)(DA)(DA)(DC)(DC)(DG)(DA) (DG)(DC) (DA) |
-Macromolecule #5: DNA (76-MER)
| Macromolecule | Name: DNA (76-MER) / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 29.101656 KDa |
| Sequence | String: (DT)(DG)(DC)(DT)(DC)(DG)(DG)(DT)(DT)(DT) (DA)(DT)(DC)(DC)(DC)(DC)(DG)(DC)(DT)(DG) (DG)(DC)(DG)(DC)(DG)(DG)(DG)(DG)(DA) (DA)(DC)(DA)(DC)(DT)(DC)(DT)(DA)(DA)(DA) (DC) (DA)(DT)(DA)(DA)(DC)(DC) ...String: (DT)(DG)(DC)(DT)(DC)(DG)(DG)(DT)(DT)(DT) (DA)(DT)(DC)(DC)(DC)(DC)(DG)(DC)(DT)(DG) (DG)(DC)(DG)(DC)(DG)(DG)(DG)(DG)(DA) (DA)(DC)(DA)(DC)(DT)(DC)(DT)(DA)(DA)(DA) (DC) (DA)(DT)(DA)(DA)(DC)(DC)(DT)(DA) (DT)(DT)(DA)(DT)(DT)(DA)(DA)(DT)(DT)(DA) (DA)(DT) (DG)(DA)(DT)(DT)(DT)(DT)(DT) (DT)(DA)(DA)(DG)(DC)(DC)(DA)(DG)(DT)(DC) (DA)(DC)(DA) (DA)(DT)(DC)(DT)(DA)(DC) (DC)(DA)(DA)(DC)(DT)(DT)(DT)(DA)(DT) |
-Macromolecule #6: DNA (61-MER)
| Macromolecule | Name: DNA (61-MER) / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 19.286447 KDa |
| Sequence | String: (DA)(DT)(DA)(DA)(DA)(DG)(DT)(DT)(DG)(DG) (DT)(DA)(DG)(DA)(DT)(DT)(DG)(DT)(DG)(DA) (DC)(DT)(DG)(DG)(DC)(DT)(DT)(DA)(DA) (DA)(DA)(DA)(DA)(DT)(DC)(DA)(DT)(DT)(DA) (DA) (DT)(DT)(DA)(DA)(DT)(DA) ...String: (DA)(DT)(DA)(DA)(DA)(DG)(DT)(DT)(DG)(DG) (DT)(DA)(DG)(DA)(DT)(DT)(DG)(DT)(DG)(DA) (DC)(DT)(DG)(DG)(DC)(DT)(DT)(DA)(DA) (DA)(DA)(DA)(DA)(DT)(DC)(DA)(DT)(DT)(DA) (DA) (DT)(DT)(DA)(DA)(DT)(DA)(DA)(DT) (DA)(DG)(DG)(DT)(DT)(DA)(DT)(DG)(DT)(DT) (DT)(DA) (DG)(DA) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 20 mM HEPES, pH 7.5, 150 mM KCl, 5 mM EDTA, 1 mM DTT, and 0.1% glycerol |
| Grid | Model: C-flat / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 15 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281.15 K / Instrument: FEI VITROBOT MARK IV |
| Details | 1uM Cas1-Cas2-DNA-IHF complexes |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 3-30 / Number grids imaged: 1 / Number real images: 3000 / Average exposure time: 6.0 sec. / Average electron dose: 1.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.6 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | model building for protein part |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-5wfe: |
-Atomic model buiding 2
| Details | model building for DNA part |
|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
| Output model |  PDB-5wfe: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)