[English] 日本語
 Yorodumi
Yorodumi- EMDB-8510: Negative-stain electron microscopy reconstructions of a dimodular... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8510 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Negative-stain electron microscopy reconstructions of a dimodular nonribosomal peptide synthetase | |||||||||
 Map data Map data | dimodular nonribosomal peptide synthetase | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Anoxybacillus kamchatkensis G10 (bacteria) Anoxybacillus kamchatkensis G10 (bacteria) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 25.8 Å | |||||||||
 Authors Authors | Haque AS / Bui KH / Schmeing TM | |||||||||
 Citation Citation |  Journal: Structure / Year: 2017 Journal: Structure / Year: 2017Title: X-Ray Crystallography and Electron Microscopy of Cross- and Multi-Module Nonribosomal Peptide Synthetase Proteins Reveal a Flexible Architecture. Authors: Michael J Tarry / Asfarul S Haque / Khanh Huy Bui / T Martin Schmeing /  Abstract: Nonribosomal peptide synthetases (NRPS) are macromolecular machines that produce peptides with diverse activities. Structural information exists for domains, didomains, and even modules, but little ...Nonribosomal peptide synthetases (NRPS) are macromolecular machines that produce peptides with diverse activities. Structural information exists for domains, didomains, and even modules, but little is known about higher-order organization. We performed a multi-technique study on constructs from the dimodular NRPS DhbF. We determined a crystal structure of a cross-module construct including the adenylation (A) and peptidyl carrier protein (PCP) domains from module 1 and the condensation domain from module 2, complexed with an adenosine-vinylsulfonamide inhibitor and an MbtH-like protein (MLP). The action of the inhibitor and the role of the MLP were investigated using adenylation reactions and isothermal titration calorimetry. In the structure, the PCP and A domains adopt a novel conformation, and noncovalent, cross-module interactions are limited. We calculated envelopes of dimodular DhbF using negative-stain electron microscopy. The data show large conformational variability between modules. Together, our results suggest that NRPSs lack a uniform, rigid supermodular architecture. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8510.map.gz emd_8510.map.gz | 9.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8510-v30.xml emd-8510-v30.xml emd-8510.xml emd-8510.xml | 22.3 KB 22.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8510_fsc_1.xml emd_8510_fsc_1.xml emd_8510_fsc_2.xml emd_8510_fsc_2.xml emd_8510_fsc_3.xml emd_8510_fsc_3.xml emd_8510_fsc_4.xml emd_8510_fsc_4.xml emd_8510_fsc_5.xml emd_8510_fsc_5.xml | 6.4 KB 6.4 KB 6.4 KB 6.4 KB 6.4 KB | Display Display Display Display Display |  FSC data file FSC data file |
| Images |  emd_8510_1.png emd_8510_1.png emd_8510_2.png emd_8510_2.png emd_8510_3.png emd_8510_3.png emd_8510_4.png emd_8510_4.png emd_8510_5.png emd_8510_5.png | 36.5 KB 42.2 KB 40.5 KB 62.8 KB 41.8 KB | ||
| Others |  emd_8510_additional_1.map.gz emd_8510_additional_1.map.gz emd_8510_additional_2.map.gz emd_8510_additional_2.map.gz emd_8510_additional_3.map.gz emd_8510_additional_3.map.gz emd_8510_additional_4.map.gz emd_8510_additional_4.map.gz | 9.7 MB 9.7 MB 9.7 MB 9.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8510 http://ftp.pdbj.org/pub/emdb/structures/EMD-8510 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8510 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8510 | HTTPS FTP |
-Validation report
| Summary document |  emd_8510_validation.pdf.gz emd_8510_validation.pdf.gz | 78.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_8510_full_validation.pdf.gz emd_8510_full_validation.pdf.gz | 77.6 KB | Display | |
| Data in XML |  emd_8510_validation.xml.gz emd_8510_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8510 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8510 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8510 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8510 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8510.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8510.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | dimodular nonribosomal peptide synthetase | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.21 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
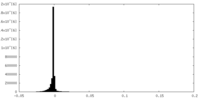
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: dimodular nonribosomal peptide synthetase, additional map #1
| File | emd_8510_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | dimodular nonribosomal peptide synthetase, additional map #1 | ||||||||||||
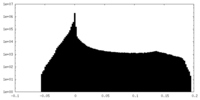
| Projections & Slices |
| ||||||||||||
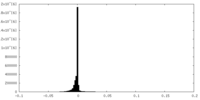
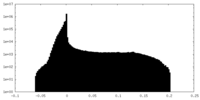
| Density Histograms |
-Additional map: dimodular nonribosomal peptide synthetase, additional map #2
| File | emd_8510_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | dimodular nonribosomal peptide synthetase, additional map #2 | ||||||||||||
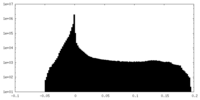
| Projections & Slices |
| ||||||||||||
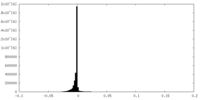
| Density Histograms |
-Additional map: dimodular nonribosomal peptide synthetase, additional map #3
| File | emd_8510_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | dimodular nonribosomal peptide synthetase, additional map #3 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: dimodular nonribosomal peptide synthetase, additional map #4
| File | emd_8510_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | dimodular nonribosomal peptide synthetase, additional map #4 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Dimodule of DhbF, a bacillibactin-synthesizing nonribosomal pepti...
| Entire | Name: Dimodule of DhbF, a bacillibactin-synthesizing nonribosomal peptide synthetase |
|---|---|
| Components |
|
-Supramolecule #1: Dimodule of DhbF, a bacillibactin-synthesizing nonribosomal pepti...
| Supramolecule | Name: Dimodule of DhbF, a bacillibactin-synthesizing nonribosomal peptide synthetase type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Anoxybacillus kamchatkensis G10 (bacteria) Anoxybacillus kamchatkensis G10 (bacteria) |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 238 KDa |
-Macromolecule #1: DhbF
| Macromolecule | Name: DhbF / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Anoxybacillus kamchatkensis G10 (bacteria) Anoxybacillus kamchatkensis G10 (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: ASMNQRLRLP LSGAQAGIWF AQQLDPANPI YNTGEYVEIH GPVDPARFEQ ALRHVLVQAE SLHAQFGED ENGPWQIIDP SPDFPFYFID VSTAANPESE ALFWMKKDLS KPVDLKCDPL F TEALFKLS DQRFFWYQRI HHIAIDGFGF SLIAQKVAET YTALMNDRMI ...String: ASMNQRLRLP LSGAQAGIWF AQQLDPANPI YNTGEYVEIH GPVDPARFEQ ALRHVLVQAE SLHAQFGED ENGPWQIIDP SPDFPFYFID VSTAANPESE ALFWMKKDLS KPVDLKCDPL F TEALFKLS DQRFFWYQRI HHIAIDGFGF SLIAQKVAET YTALMNDRMI AADEAFASFR EV IEEEKVY HASEQYERDR QFWLERFRDQ PEAVSLSDRA ARASHTFIRK TVHLSVAQTE RLK QSTQYW KAGWHELFLA ATALYLHRLT SATDVILGLP MMNRLGSVAL NVPAMVMNLV PFRL HLHAD MKMSQLLSAV RQEIKEIKQH HKYRHEQLRR DLKLLGENQR LFGVQVNIMP FDYGL HFDG YQGITHNLSA GPVDDLAINV YDRTDGNGLR IDFDANPEIY QADDLAIHQS RFLQIL EML TVLQEDVTIG SVELLLEQER RQVLEIWNET ARHHSEASFL QLFEKQAQQH PEAMAVI CE NQMLRYQELN EQANRLAHLL VEKGAGPEQY VALALPRSAD MVIAMLAVLK AGAAYLPI D PEYPKERLAF MLDDAEPLCI ITNSATQPKL SAISPLSMIV LDHPETEEAI KRYPSTNVE NGRSALHPAY VIYTSGSTGK PKGVVVPFQN LNNFLFAMQD KFMLNERDRW LAVTTIAFDI AALEIFLPL MSGARLIVAK KEAIHDPQTL AALISNEEIT IMQATPTLWH MLVTYHPDSI A GLRVLVGG EALPSSLASA LDQLGCEITN LYGPTETTIW STMATLQQDD IVAPVIGKPI WN TQVYVLD RHLQPVPPGV AGELYIAGEG VARGYLKRPD LTAERFVANP YGPPGSRMYR TGD LVRWRK DGSLDYIGRV DYQVKLRGFR IEIGEIEAIL TKYDEVERAV VVAREDQPGA PRLV AYLIP RGSKGALDLA ELRLYVSEKL PDYMVPSAFM ILEEFPLTPN GKIDRKALPV PDWTV TMKG RKPRTPQEEI LCELFAEVLD LSAVGIDDNF FELGGHSLLA ARLISRIRDV LGVELA IGK LFESPTVASL VHHLKEAAHG KPPVKAYAHK DEIPLSFAQR RLWFLHHLEG PSPTYNI PV VVHLTGALQI AALRQALYDV VERHETLRTI FPDRSGTSRQ IVLEPHQARP ELIVQEIS E NELSRVLNEA VRYSFDLAKE PPIRAQLFVL GENRYVLLLL MHHIAADGWS LTPLTRDLA SAYQAHCQQQ KVDWPPLPVT YADYALWQQE FLGNENHPDH LIAKQLDYWK QTLADLPEEL EWSTDYPRP AESSYEGGVV EFKLDAELHQ RLLALARENK TSLFMVLQAG FAALLTRLGA G TDIPIGSP IAGRNDDALE DLVGMFINTL VLRMDTSGNP SFRELLARVK QVNLSAYENQ DL PFERLVE VLNPVRSRAK HPLFQVMFVF QNTPEPKLEL QGLESRLEVR SVGSAKFDLT LEL RENRMG DGSPAGLIGL FEYSRDLFQQ ETVERFAKRL CQLLKEVVTN PELPIGQIHM LLPE ERKLL LNQAEHRQRH LSAETLPALF EKQVQRVPDA TAVVFEDQKL SYAELNKKAN QLAHF LISK GVGPERVVAL ALPRSLEMVI GILAVLKAGG AYLPLDPNYP EDRIAYMMED AQPMYV IAN QQMAVKLPHT PNVEHIVVDA EHVVGQIGHY PETNPNDADR IEALSPYHMA YIIYTSG ST GKPKGVMIPH QNVVRLFRST EHWFQFSEND SWTLFHSYAF DFSVWEIWGP FLYGGRLV V VPHHVSRSPK EFLQLLVEQN VTVLNQTPSA FYQLMQADRE NSEWSKQLSL RFVIFGGEA LELSRLEDWY ERHPENQPKL INMYGITETT VHVSYIELDR TILSLKGNSL IGCSIPDLEV YVLDDQLQP VPPGVIGEMY VAGAGLARGY LGRPDLTAER FIANPFGPPG SRMYRTGDLA K WRVDGTLD YIGRADHQVK IRGYRIELGE IEAVLAKHPQ IAQVAVIVRE DQPGDKRLVA YI VPAKDAV FESSELRKYV AASLPDYMIP SAFVMIEALP LTPNGKLDRK ALPMPDLAIE VNG RGPRTP QEEMLCDLFM EILRLPRVGI DDGFFELGGH SLLAVQLMSR IRETFGVELS IGDL FEAPT VAGLAEKLEM GSS |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Staining | Type: NEGATIVE / Material: Uranyl formate | ||||||||||||
| Grid | Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 7.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Average electron dose: 12.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)